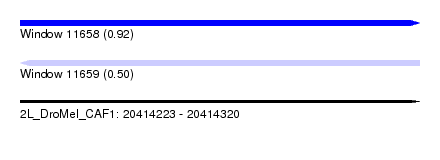
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,414,223 – 20,414,320 |
| Length | 97 |
| Max. P | 0.919688 |

| Location | 20,414,223 – 20,414,320 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.92 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
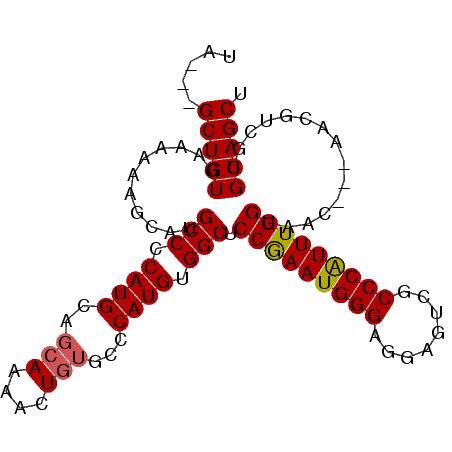
>2L_DroMel_CAF1 20414223 97 + 22407834 UA---GCUGUGAAAAAAGCGUGCCCCAUGCAGCAGAACUGUGCCCAUGUGGCUCCGAAUGGGAGGAGUCGCCCAUUUGGUAACGGUAACGUCGUGCAGCU .(---((((((......(((((((.((((..(((....)))...)))).))).(((((((((........)))))))))........))))..))))))) ( -34.10) >DroSec_CAF1 8836 97 + 1 UA---GCUGUGAAAAAAGCAUGCCCCAUGCAGCAGAACUGUGCCCAUGUGGCUCCGAAUGGGAGGAGUCGCCCAUUUGGUAACGGUAACGUCGUGCAGCU .(---(((((.......(((((...)))))......(((((..(((.((((((((........)))))))).....)))..)))))........)))))) ( -33.80) >DroSim_CAF1 9970 97 + 1 UA---GCUGUGAAAAAAGCAUGCCCCAUGCAGCAGAACUGUGCCCAUGUGGCUCCGAAUGGGAGGAGUCGCCCAUUUGGUAACGGUAACGUCGUGCAGCU .(---(((((.......(((((...)))))......(((((..(((.((((((((........)))))))).....)))..)))))........)))))) ( -33.80) >DroEre_CAF1 8861 94 + 1 UAGUCGCUGUGAAAAAAGCAUGCCCCAUGCAGCAAAACUGUGCCCAUGUGGCUCCGAAUGGGAGGAGUCGCCCAUUUGGU------AACGUCGUGCAGCU .....(((((...........(((.((((..(((......))).)))).))).(((((((((........))))))))).------........))))). ( -31.40) >DroYak_CAF1 8512 91 + 1 UA---GCUGUGAAAAAAGCAUGCCCCAUGCAGCAAAACUGUGCCCAUGUGGCUCCAAAUGGGAGGAGUCGCCCAUUUGGU------AACGUCGUGCAGCU .(---(((((...........(((.((((..(((......))).)))).))).(((((((((........))))))))).------........)))))) ( -32.00) >DroAna_CAF1 7372 79 + 1 GA---GCUGUGAAAAA----UGCCCCAUGCA--AAAACUGUGCCCAUGUGGCUCCAAAUGGG---AGUCACCCGUUUGGUAAC---------GUGCAGCU .(---(((((......----.(((.((((((--.......)))..))).))).(((((((((---.....)))))))))....---------..)))))) ( -27.90) >consensus UA___GCUGUGAAAAAAGCAUGCCCCAUGCAGCAAAACUGUGCCCAUGUGGCUCCGAAUGGGAGGAGUCGCCCAUUUGGUAAC___AACGUCGUGCAGCU .....(((((...........(((.((((..(((....)))...)))).))).(((((((((........)))))))))...............))))). (-29.94 = -29.92 + -0.03)

| Location | 20,414,223 – 20,414,320 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.48 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
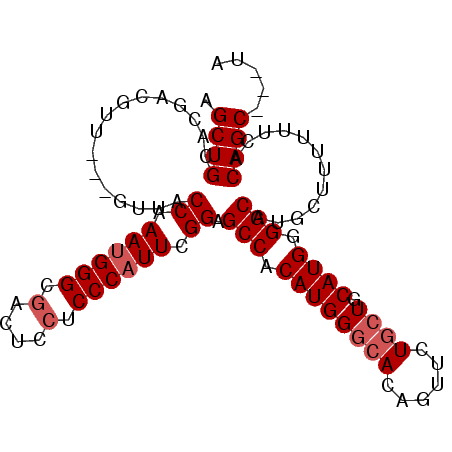
>2L_DroMel_CAF1 20414223 97 - 22407834 AGCUGCACGACGUUACCGUUACCAAAUGGGCGACUCCUCCCAUUCGGAGCCACAUGGGCACAGUUCUGCUGCAUGGGGCACGCUUUUUUCACAGC---UA (((((...((((....)))).((.((((((.(....).)))))).)).(((.((((((((......)))).)))).)))............))))---). ( -31.60) >DroSec_CAF1 8836 97 - 1 AGCUGCACGACGUUACCGUUACCAAAUGGGCGACUCCUCCCAUUCGGAGCCACAUGGGCACAGUUCUGCUGCAUGGGGCAUGCUUUUUUCACAGC---UA (((((...((((....)))).((.((((((.(....).)))))).)).(((.((((((((......)))).)))).)))............))))---). ( -31.60) >DroSim_CAF1 9970 97 - 1 AGCUGCACGACGUUACCGUUACCAAAUGGGCGACUCCUCCCAUUCGGAGCCACAUGGGCACAGUUCUGCUGCAUGGGGCAUGCUUUUUUCACAGC---UA (((((...((((....)))).((.((((((.(....).)))))).)).(((.((((((((......)))).)))).)))............))))---). ( -31.60) >DroEre_CAF1 8861 94 - 1 AGCUGCACGACGUU------ACCAAAUGGGCGACUCCUCCCAUUCGGAGCCACAUGGGCACAGUUUUGCUGCAUGGGGCAUGCUUUUUUCACAGCGACUA .((((.........------.((.((((((.(....).)))))).)).(((.((((((((......)))).)))).)))............))))..... ( -29.80) >DroYak_CAF1 8512 91 - 1 AGCUGCACGACGUU------ACCAAAUGGGCGACUCCUCCCAUUUGGAGCCACAUGGGCACAGUUUUGCUGCAUGGGGCAUGCUUUUUUCACAGC---UA (((((.........------.(((((((((.(....).))))))))).(((.((((((((......)))).)))).)))............))))---). ( -34.30) >DroAna_CAF1 7372 79 - 1 AGCUGCAC---------GUUACCAAACGGGUGACU---CCCAUUUGGAGCCACAUGGGCACAGUUUU--UGCAUGGGGCA----UUUUUCACAGC---UC (((((...---------....(((((.(((.....---))).))))).(((.((((.(.........--).)))).))).----.......))))---). ( -24.10) >consensus AGCUGCACGACGUU___GUUACCAAAUGGGCGACUCCUCCCAUUCGGAGCCACAUGGGCACAGUUCUGCUGCAUGGGGCAUGCUUUUUUCACAGC___UA .((((................((.((((((.(....).)))))).)).(((.((((((((......)))).)))).)))............))))..... (-25.48 = -26.15 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:20 2006