
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,409,370 – 20,409,569 |
| Length | 199 |
| Max. P | 0.990492 |

| Location | 20,409,370 – 20,409,490 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20409370 120 + 22407834 UAAAUGGAGACAAAGUUUACAUGUUAACAAUAUUAUCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUUCGAUCGAAUCAUCCGAAGCAGUCUUAUAUUUCCUUGGAUGAUUUCUU ....(((((.((..((..(((((..(((.........)))..)))))..))..)))))))..................(((((((((((.(.(((.....))).).)))))))))))... ( -27.20) >DroSec_CAF1 4091 120 + 1 UAAAUGGAGACAAAGUUUACAUGUUAACAAUGCUAUCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUCGAAUCAUCCGAAGCAUUUUCAUAUUACCUUGGAUGAUUUCUU ....(((((.((..((..(((((..(((.........)))..)))))..))..)))))))..................(((((((((((.................)))))))))))... ( -26.43) >DroSim_CAF1 5063 120 + 1 UAAAUGGAGACAAAGUUUACAUGUUAAUAAUGCUAUCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUAGAAUCAUCCGAAGCAGUCUUAUAUUUCCUUGGAUGAUUUCUU ....(((((.((..((..(((((..(((.........)))..)))))..))..)))))))..................(((((((((((.(.(((.....))).).)))))))))))... ( -25.20) >DroEre_CAF1 4108 120 + 1 UAAAUGGAGACAAAGUUUACAUGUUAGCAGUAUUUUCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAUGUAUCCGAUCCAAUCAUCCGAAGCAGUCUUAUAUUACCUUGGAUGAUUUCUU ....(((((.((..((..(((((..(((.........)))..)))))..))..)))))))...................((((((((((.(.(((.....))).).)))))))))).... ( -25.90) >consensus UAAAUGGAGACAAAGUUUACAUGUUAACAAUACUAUCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUCGAAUCAUCCGAAGCAGUCUUAUAUUACCUUGGAUGAUUUCUU ....(((((.((..((..(((((..(((.........)))..)))))..))..)))))))..................(((((((((((.(.(((.....))).).)))))))))))... (-25.85 = -25.98 + 0.12)

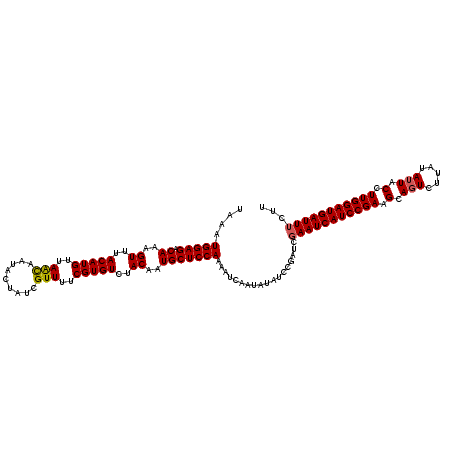
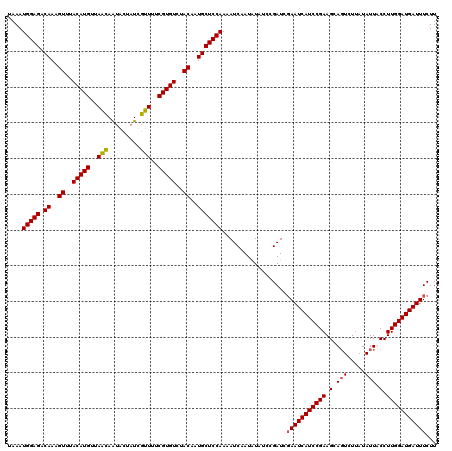
| Location | 20,409,410 – 20,409,530 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20409410 120 + 22407834 UUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUUCGAUCGAAUCAUCCGAAGCAGUCUUAUAUUUCCUUGGAUGAUUUCUUUGGUGCACAGUGCACAGCUAAUGUAAUCCCAAUGGAGUUA ..............((((((.......(((((((((..(((((((((((.(.(((.....))).).)))))))))))..)))((((.....))))....)))))).......)))))).. ( -28.34) >DroSec_CAF1 4131 120 + 1 UUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUCGAAUCAUCCGAAGCAUUUUCAUAUUACCUUGGAUGAUUUCUUUGGUGCAUAGUGCACAGCUAAUGUAAUCCCAAUGGAGUUA ..............((((((............((((..(((((((((((.................)))))))))))..))))((((((((.....))).))))).......)))))).. ( -28.43) >DroSim_CAF1 5103 120 + 1 UUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUAGAAUCAUCCGAAGCAGUCUUAUAUUUCCUUGGAUGAUUUCUUUGGUGCACAGUGCACAGCUAAUGUAAUCCCAAUGGAGUUA ..............((((((.............(((..(((((((((((.(.(((.....))).).)))))))))))..)))((((.....)))).................)))))).. ( -28.40) >DroEre_CAF1 4148 118 + 1 UUCGUGUCUACAAUGCUCCAAAAUCAAUGUAUCCGAUCCAAUCAUCCGAAGCAGUCUUAUAUUACCUUGGAUGAUUUCUUUGGUGCACAG--CAAGGCAUAAAUUUUUCUUAUGGAGUUA ..............((((((.......((((.((((...((((((((((.(.(((.....))).).))))))))))...))))))))..(--(...))..............)))))).. ( -25.60) >consensus UUCGUGUCUACAAUGCUCCAAAAUCAAUAUAUCCGAUCGAAUCAUCCGAAGCAGUCUUAUAUUACCUUGGAUGAUUUCUUUGGUGCACAGUGCACAGCUAAUGUAAUCCCAAUGGAGUUA ..............((((((.............(((..(((((((((((.(.(((.....))).).)))))))))))..)))((((.....)))).................)))))).. (-23.52 = -24.52 + 1.00)

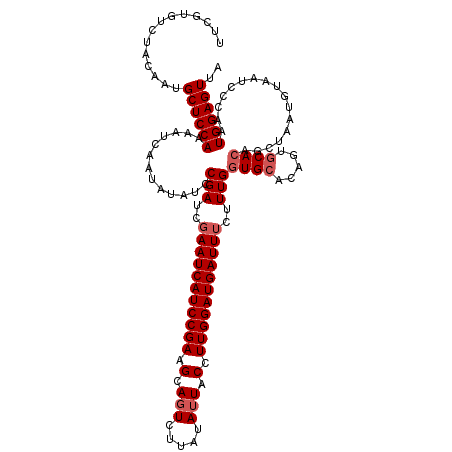

| Location | 20,409,450 – 20,409,569 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.84 |
| Energy contribution | -21.65 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20409450 119 - 22407834 AGACACAAAUUGAAGGAAGGUAUACAAAAUAUGCCACUUUAACUCCAUUGGGAUUACAUUAGCUGUGCACUGUGCACCAAAGAAAUCAUCCAAGGAAAUAUAAGACUGCUUCGGAUGAU .........((((((...((((((.....)))))).)))))).(((....)))...........((((.....)))).......((((((((((.(..........).))).))))))) ( -23.50) >DroSec_CAF1 4171 119 - 1 AGACACAGAUUAAAGGAAGGUAUAUAAAAUAUGCCACUUUAACUCCAUUGGGAUUACAUUAGCUGUGCACUAUGCACCAAAGAAAUCAUCCAAGGUAAUAUGAAAAUGCUUCGGAUGAU .(.(((((.((((((...(((((((...))))))).)))))).(((....))).........))))))................(((((((.(((((.........))))).))))))) ( -28.50) >DroSim_CAF1 5143 119 - 1 AGACACAGAUUAAAGGAAGGUAUAUAAAAUAUGCCACUUUAACUCCAUUGGGAUUACAUUAGCUGUGCACUGUGCACCAAAGAAAUCAUCCAAGGAAAUAUAAGACUGCUUCGGAUGAU .(.(((((.((((((...(((((((...))))))).)))))).(((....)))................)))))).........((((((((((.(..........).))).))))))) ( -26.10) >DroEre_CAF1 4188 116 - 1 AGACACAGAUUAAAGGUAAGUAC-UAAUACAUGCCACUUUAACUCCAUAAGAAAAAUUUAUGCCUUG--CUGUGCACCAAAGAAAUCAUCCAAGGUAAUAUAAGACUGCUUCGGAUGAU .(.(((((......((((.(((.-...))).))))..........((((((.....)))))).....--)))))).........(((((((.(((((.........))))).))))))) ( -24.60) >consensus AGACACAGAUUAAAGGAAGGUAUAUAAAAUAUGCCACUUUAACUCCAUUGGGAUUACAUUAGCUGUGCACUGUGCACCAAAGAAAUCAUCCAAGGAAAUAUAAGACUGCUUCGGAUGAU .(.(((((.((((((...((((((.....)))))).)))))).(((....)))................)))))).........(((((((.(((((.........))))).))))))) (-19.84 = -21.65 + 1.81)

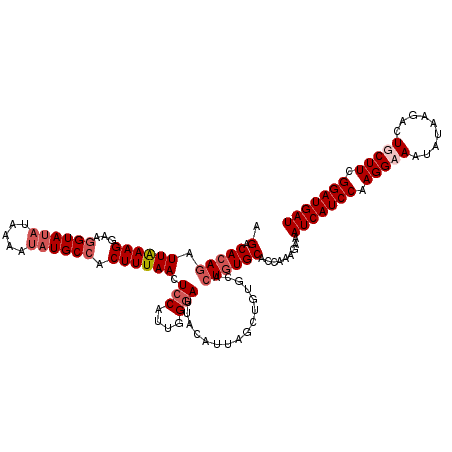
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:19 2006