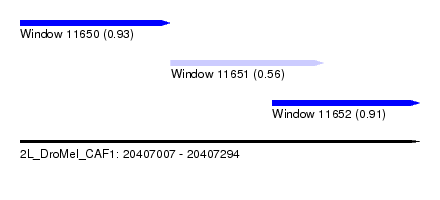
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,407,007 – 20,407,294 |
| Length | 287 |
| Max. P | 0.926242 |

| Location | 20,407,007 – 20,407,115 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.91 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -12.78 |
| Energy contribution | -12.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20407007 108 + 22407834 ACAUUCGUUCACAAUACAUUCGUUGUUCAUAUGAAA----AUGGA--UUGGAGUCAAAGUGGCACACUUAA----UCCAUUUUUAUUUGUGUCUCAAAUGCUCCAAUAUCAAAUGCAA ...(((((..(((((......)))))....))))).----.((((--(((((((....(.((((((..(((----.......)))..)))))).)....)))))))).)))....... ( -24.30) >DroSec_CAF1 1663 99 + 1 ACAUUCGUUCACAU---------CGCUCAUAUGGAA----AUGGA--UUGGAGUCAAAGUGGCACACUUAA----ACACUUUUUAUUUGUGUCUCAAAUGCUCCAAUAUCAAAUGCAA ....(((((..(((---------.......)))..)----))))(--(((((((....(.((((((..(((----(.....))))..)))))).)....))))))))........... ( -22.30) >DroSim_CAF1 2658 99 + 1 ACAUUCGUUCACAA---------UGCUCAUAUGGAA----AUGGA--UUGGAGUCAAAGUGGCACACUUAA----UCAGUUUUUAUUUGUGUCUCAAAUGCUCCAAUAUCAAAUGCAA ..............---------(((((((......----))))(--(((((((....(.((((((..(((----.......)))..)))))).)....)))))))).......))). ( -21.60) >DroEre_CAF1 1621 102 + 1 ACUUUCGUGUACAUUA------UUGCAUAUAUCGU--------CA--UUGGAGUCAAAGUGGCAUUCUUCAAAAAUAAUAUUUGAUCUGUGUCUCAAAUGCUCCAACAUAAACUAGAA ...((((((((.....------.))))).......--------..--(((((((....(.(((((...(((((.......)))))...))))).)....))))))).........))) ( -19.70) >DroYak_CAF1 1668 101 + 1 ACUUUCGUCUACAUUA------UUGCAUAUAUCGU--------CA--UUGGAGUCAAAGUGGCAUUCUUCAAAAAUAAUAUUCGAUCUGUGUCUCAAAAGCUCCAACAUAAAC-AGAU ......((((.((...------.))..........--------..--(((((((....(.(((((...((.............))...))))).)....))))))).......-)))) ( -13.62) >DroAna_CAF1 1638 109 + 1 GUGUUUGAAUACAUUA------U---CCUUAAGGUAUAUCCUGUAUCCUGGAGUCAAAGUGGCAUUUAUGGGAACUCUUAUCCGAUUUGUGUCUCAAAUACUCCAAAACAAACUAGUG ..(((((.(((((.((------(---((....)).)))...)))))..((((((....(.(((((..((.(((.......))).))..))))).)....))))))...)))))..... ( -26.30) >consensus ACAUUCGUUCACAUUA______UUGCUCAUAUGGAA____AUGGA__UUGGAGUCAAAGUGGCACACUUAA____UCAUAUUUGAUUUGUGUCUCAAAUGCUCCAAUAUAAAAUACAA ...............................................(((((((....(.(((((...(((...........)))...))))).)....)))))))............ (-12.78 = -12.33 + -0.44)

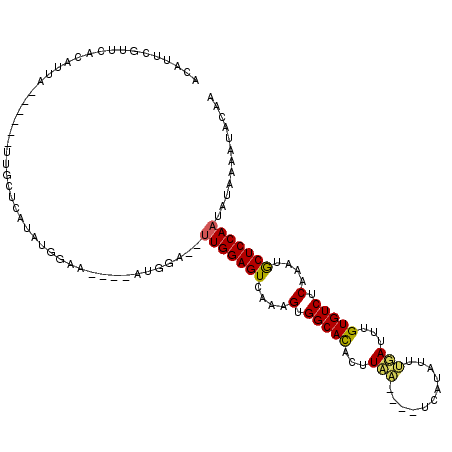
| Location | 20,407,115 – 20,407,225 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.36 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -14.70 |
| Energy contribution | -13.98 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20407115 110 + 22407834 AGGCGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGCCUACAAUAGCUCCUGA-----UAGCUUUACAUUCCUUUAUAAGACUCCCGUCUUCGGAGA ((((((((((((((.((.(((((.......)))))..)))))).))...)))))))).....((((.....-----.)))).....((((.....(((((....))))).)))). ( -24.60) >DroSec_CAF1 1762 110 + 1 AGACGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGUCUACAAUAGCUCCUGA-----UAGCUUUACACUCCUUCAUAGAACUCCAGUCCCCGGAGA ((((((((((((((.((.(((((.......)))))..)))))).))...)))))))).....((((.....-----.))))..................((((.......)))). ( -21.90) >DroSim_CAF1 2757 110 + 1 AGACGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGUCUACAAUACCUUCUGA-----UAGCUUUACACUCCUUCUUAGAACUCCAGUCCCCGGAGA ((((((((((((((.((.(((((.......)))))..)))))).))...))))))))........((((((-----.................))))))((((.......)))). ( -20.93) >DroEre_CAF1 1723 98 + 1 GGUCGAACGGCAUCCUGUAUAAUCCU-CAUAUUAUAGCAGAUGACUUAAGUUGGACUACAAUAGUAUUAAAUGAUUCAGGUCACUAAUUC----------------CCUGGGAGA ((((.(((((((((.((((((((...-...))))).))))))).))...))).)))).................((((((..........----------------))))))... ( -18.30) >DroYak_CAF1 1769 98 + 1 AGUCGAACGGCAUCCUGGAUAAUCCU-CAUAUUAUAGCAGAUGACUUGUGUUCGACUACAAUAUUAUUAAAUGAUAUAGCUCAAUAAUUC----------------CUCGGGAGA ((((((((((((((.((.(((((...-...)))))..)))))).))...))))))))...(((((((...)))))))..........(((----------------(...)))). ( -18.60) >DroAna_CAF1 1747 91 + 1 AGACGGACGGCAUCCUGAAUAAUGUAUCCUAUUAUAUCAGAUGACUUGUGUUCGUCUACAUU---------------------CUACCCCUUUUAAGAUCUC---UUUGUGGAGA ((((((((((((((.(((((((((.....)))))).))))))).))...)))))))).....---------------------...............((((---(....))))) ( -21.90) >consensus AGACGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGUCUACAAUAGCUUCUGA_____UAGCUUUACACUCCUU__UAGAACUC___UCCCCGGAGA ((((((((((((((....(((((.......)))))....)))).))...)))))))).......................................................... (-14.70 = -13.98 + -0.72)

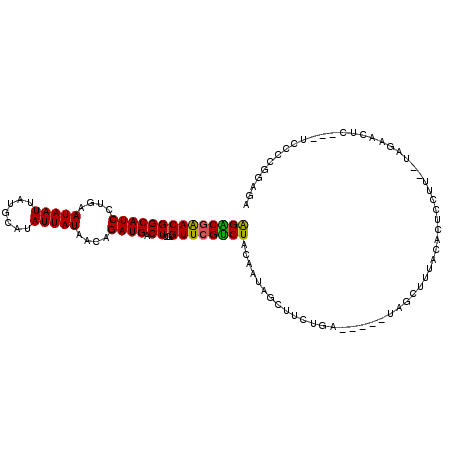
| Location | 20,407,188 – 20,407,294 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.95 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.05 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20407188 106 + 22407834 GCUUUACAUUCCUUUAUAAGACUCCCGUCUUCGGAGACAAAGUUUACAUGCUAAGGA---UAAG--AUCUUUUUCGUGUCUACAAUGCUCCAAGAGCAAAUGU----UCGUUUAA (((((............(((((....))))).((((.((..((..(((((..(((((---(...--))))))..)))))..))..)))))).)))))......----........ ( -25.20) >DroSec_CAF1 1835 106 + 1 GCUUUACACUCCUUCAUAGAACUCCAGUCCCCGGAGACAAAGUUUACAUGCUAAGGA---UAAG--AUCUUUUUCGUGUCUACAAUGCUCCAAGAGCAAAUGA----UCGUUUAA (((((......((....)).............((((.((..((..(((((..(((((---(...--))))))..)))))..))..)))))).)))))......----........ ( -20.70) >DroSim_CAF1 2830 106 + 1 GCUUUACACUCCUUCUUAGAACUCCAGUCCCCGGAGACAAAGUUUACAUGCUAAGGA---UAAG--AUCUUUUUCGUGUCUACAAUGCUCCAAGAGCAAAUGU----UCGUUCAA ..................((((..........((((.((..((..(((((..(((((---(...--))))))..)))))..))..))))))..((((....))----)))))).. ( -24.90) >DroEre_CAF1 1800 95 + 1 GGUCACUAAUUC----------------CCUGGGAGACAAAGUUUACAUGUAAAAGAUUAUGUGGCUUCUUUUUCGUGUCUACAAUGCUCCAAAAUAAACUGU----UCGUUUAA ............----------------....((((.((..((..(((((.((((((..........)))))).)))))..))..))))))....(((((...----..))))). ( -19.30) >DroYak_CAF1 1846 92 + 1 GCUCAAUAAUUC----------------CUCGGGAGACAAAGUUUACAUGUUAAGGA---UGUGGCUUCUUUUUCGUGUCUACAAUGCUCCAACAUAAACUGU----UCGUUUAA ............----------------....((((.((..((..(((((..(((((---.......)))))..)))))..))..))))))....(((((...----..))))). ( -19.50) >DroAna_CAF1 1809 105 + 1 -----CUACCCCUUUUAAGAUCUC---UUUGUGGAGACAAAGUUUACAUGUUGAAGUCCUUACA--AUCUUCCUCGUGUCUACAACGCUCCUAGAACAGCCCCCAACUCGAACGG -----.....((.((..((.((((---(....)))))....((..(((((..((((........--..))))..)))))..))...(((........)))......))..)).)) ( -18.00) >consensus GCUUUACACUCCUU__UAGAACUC___UCCCCGGAGACAAAGUUUACAUGCUAAGGA___UAAG__AUCUUUUUCGUGUCUACAAUGCUCCAAGAGCAAAUGU____UCGUUUAA ................................((((.((..((..(((((..(((((..........)))))..)))))..))..))))))........................ (-15.22 = -15.05 + -0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:14 2006