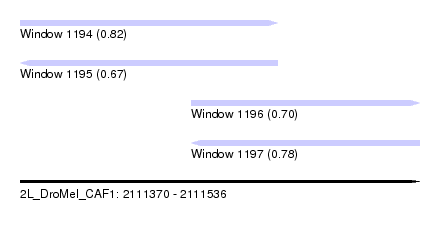
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,111,370 – 2,111,536 |
| Length | 166 |
| Max. P | 0.818316 |

| Location | 2,111,370 – 2,111,477 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -22.26 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
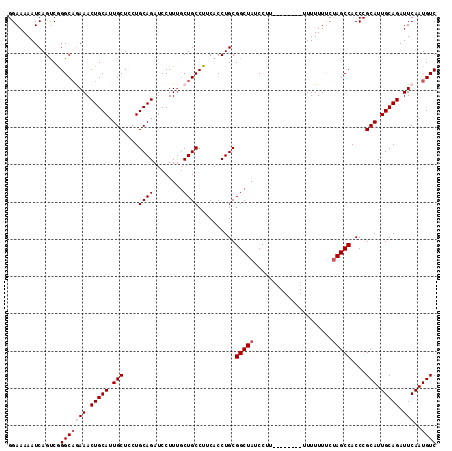
>2L_DroMel_CAF1 2111370 107 + 22407834 GGAAAAAUCAGUCGGGCUGAAACUGCAUUGCUCCUGCAGAUCCUUUGCUGCCUUCACCUGCGGCUAUCCUU--------UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUC .(((...((((.....))))..(((((.(((....((((........))))..........(((((.....--------........)))))....))).))))).)))...... ( -27.02) >DroSec_CAF1 119302 106 + 1 GGAAAAAUCAGUCGGGCAGAAACUGCAUUGCUCCUGCAGAUCCUUUGCUGCCUUCACCUGCGGCUAUCCU---------UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUC ..............(((((((.(((((.(((....((((........))))..........(((((....---------........)))))....))).))))).)))..)))) ( -25.70) >DroSim_CAF1 119112 107 + 1 GGAAAAAUCAGUCGGGCAGAAACUGCAAUGCUCCUGCAGAUCCUUUGCUGCCUUCACCUGCGGCUAUCCUC--------UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUC ..............(((((((.(((((((((....((((........))))..........(((((.....--------........)))))....))))))))).)))..)))) ( -28.82) >DroEre_CAF1 121985 114 + 1 GGAAAAAUCAGUCAGGCAAAAACUGCAUUGCUCCUGCAGAUCCUUU-CUGCCUUCACCUGCGGCUAUCCUUUUAUUUCUUUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUC ..............((((....(((((.(((....(((((.....)-))))..........(((((.....................)))))....))).)))))......)))) ( -24.40) >DroYak_CAF1 124264 106 + 1 GGAAAAAUCAGCCAGGCAAAAACUGCAUUGCUCCUGCAGAUCCUUCGCUGCCUCCACCUGCGGCUAUCCUU---------UUUUUCCCAGCCACCCGCAUUGCAGAUUCAAUGUC (((((((......(((......(((((.......))))).......(((((........)))))...))).---------))))))).........((((((......)))))). ( -22.40) >consensus GGAAAAAUCAGUCGGGCAGAAACUGCAUUGCUCCUGCAGAUCCUUUGCUGCCUUCACCUGCGGCUAUCCUU________UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUC ..............(((((((.(((((.(((....((((........))))..........(((((.....................)))))....))).))))).)))..)))) (-22.26 = -23.06 + 0.80)

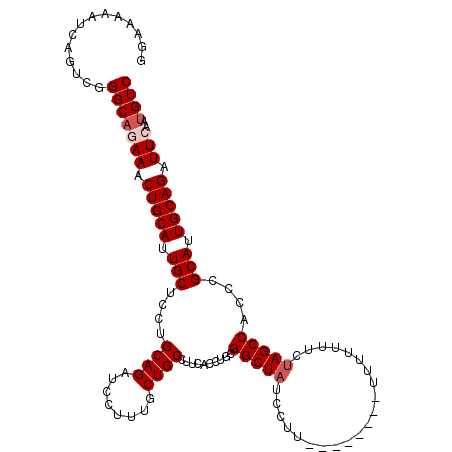
| Location | 2,111,370 – 2,111,477 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -27.30 |
| Energy contribution | -26.90 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2111370 107 - 22407834 GACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA--------AAGGAUAGCCGCAGGUGAAGGCAGCAAAGGAUCUGCAGGAGCAAUGCAGUUUCAGCCCGACUGAUUUUUCC ....(((((.(((((.(((..(((((((........--------.....)))))))........((((........))))....))).))))).)))))................ ( -30.32) >DroSec_CAF1 119302 106 - 1 GACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA---------AGGAUAGCCGCAGGUGAAGGCAGCAAAGGAUCUGCAGGAGCAAUGCAGUUUCUGCCCGACUGAUUUUUCC (.(((((......))))))..(((((((........---------....))))))).((.((((.(((....((....((((((((......))))))))))..))).)))).)) ( -29.20) >DroSim_CAF1 119112 107 - 1 GACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA--------GAGGAUAGCCGCAGGUGAAGGCAGCAAAGGAUCUGCAGGAGCAUUGCAGUUUCUGCCCGACUGAUUUUUCC ......(((.(((((((((..(((((((........--------.....)))))))........((((........))))....))))))))).))).................. ( -32.22) >DroEre_CAF1 121985 114 - 1 GACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAAAGAAAUAAAAGGAUAGCCGCAGGUGAAGGCAG-AAAGGAUCUGCAGGAGCAAUGCAGUUUUUGCCUGACUGAUUUUUCC (.(((((......))))))(((((((((.....................))))))).(((..((((((-(((....(((((.......)))))))))))))).))).......)) ( -31.20) >DroYak_CAF1 124264 106 - 1 GACAUUGAAUCUGCAAUGCGGGUGGCUGGGAAAAA---------AAGGAUAGCCGCAGGUGGAGGCAGCGAAGGAUCUGCAGGAGCAAUGCAGUUUUUGCCUGGCUGAUUUUUCC ..........((((..(((..(((((((.......---------.....)))))))..)))...)))).((((((((.((.((.((((........)))))).)).)))))))). ( -33.10) >consensus GACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA________AAGGAUAGCCGCAGGUGAAGGCAGCAAAGGAUCUGCAGGAGCAAUGCAGUUUCUGCCCGACUGAUUUUUCC ......(((.(((((.(((..(((((((.....................)))))))........((((........))))....))).))))).))).................. (-27.30 = -26.90 + -0.40)

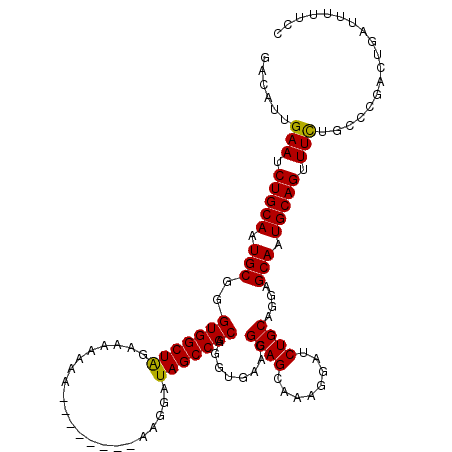
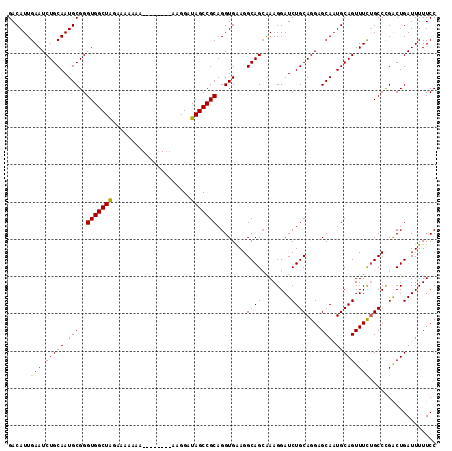
| Location | 2,111,441 – 2,111,536 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -12.50 |
| Consensus MFE | -10.82 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2111441 95 + 22407834 ---UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACAAAAAACGGAACAAAAACAC ---..(((((((.((.....)).((((........((((((....))))))...((.....)).))))..)))))))..................... ( -12.50) >DroSec_CAF1 119372 95 + 1 ---UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACAAAGAACGGAACAAAAACAC ---..(((((((.((.....)).((((........((((((....))))))...((.....)).))))..)))))))..................... ( -12.50) >DroSim_CAF1 119183 95 + 1 ---UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACAAAGAACGGAACAAAAACAC ---..(((((((.((.....)).((((........((((((....))))))...((.....)).))))..)))))))..................... ( -12.50) >DroEre_CAF1 122060 98 + 1 UCUUUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACACGGAACGGAACAAAAACAC (((..(((((((.((.....)).((((........((((((....))))))...((.....)).))))..)))))))...)))............... ( -13.40) >DroYak_CAF1 124335 94 + 1 ----UUUUUCCCAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACAAAGAACGGAACAAAACCAG ----...((((..((.....)).((((........((((((....))))))...((.....)).)))).................))))......... ( -11.60) >consensus ___UUUUUUUCUAGCCACCCGCAUUGCAGAUUCAAUGUCAAAAAUUUGACAAACUCAACAAGAAGCGAAAAGAAAAACAAAGAACGGAACAAAAACAC .....(((((((.((.....)).((((........((((((....))))))...((.....)).))))..)))))))..................... (-10.82 = -11.22 + 0.40)

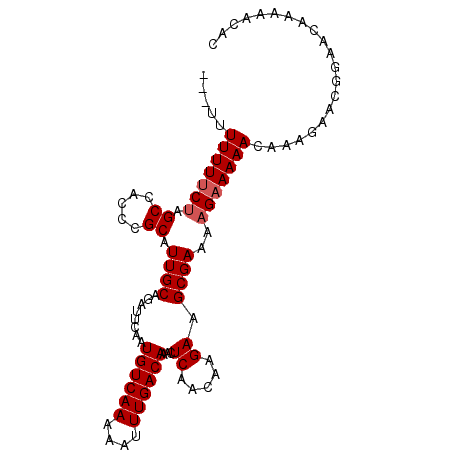
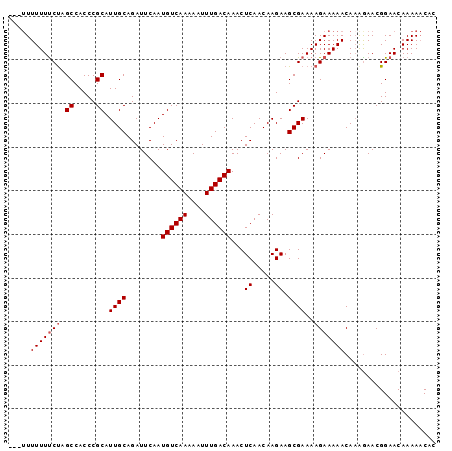
| Location | 2,111,441 – 2,111,536 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.12 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2111441 95 - 22407834 GUGUUUUUGUUCCGUUUUUUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA--- .....................(((((((..(((((((((((.((...((((((....)))))).....)).)))).).))))))..)))))))..--- ( -19.30) >DroSec_CAF1 119372 95 - 1 GUGUUUUUGUUCCGUUCUUUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA--- .....................(((((((..(((((((((((.((...((((((....)))))).....)).)))).).))))))..)))))))..--- ( -19.30) >DroSim_CAF1 119183 95 - 1 GUGUUUUUGUUCCGUUCUUUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA--- .....................(((((((..(((((((((((.((...((((((....)))))).....)).)))).).))))))..)))))))..--- ( -19.30) >DroEre_CAF1 122060 98 - 1 GUGUUUUUGUUCCGUUCCGUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAAAGA .................(...(((((((..(((((((((((.((...((((((....)))))).....)).)))).).))))))..)))))))...). ( -19.70) >DroYak_CAF1 124335 94 - 1 CUGGUUUUGUUCCGUUCUUUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUGGGAAAAA---- (..(((....(((((...((((..(((.....((((......)))).((((((....))))))..)))...)))).))))).)))..)......---- ( -20.40) >consensus GUGUUUUUGUUCCGUUCUUUGUUUUUCUUUUCGCUUCUUGUUGAGUUUGUCAAAUUUUUGACAUUGAAUCUGCAAUGCGGGUGGCUAGAAAAAAA___ .....................(((((((..(((((((((((.((...((((((....)))))).....)).)))).).))))))..)))))))..... (-19.52 = -19.20 + -0.32)

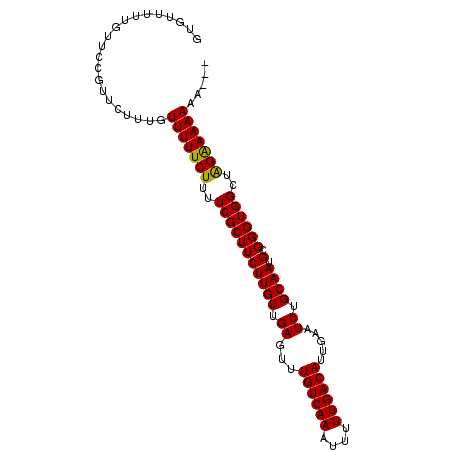
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:06 2006