
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,364,815 – 20,364,914 |
| Length | 99 |
| Max. P | 0.940296 |

| Location | 20,364,815 – 20,364,914 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -26.18 |
| Energy contribution | -26.41 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20364815 99 - 22407834 CUCAGCUAUGCCCACUUGUCUUGCUGAAUG--U-CAAAGUUUUGAGAGGAAGAAAACUUC----AGCAUUUAGCUGAAGAUGACUGCGCAUAGCCACGCCCCCAUC ....(((((((.((.(..((((((((((((--(-..(((((((.........))))))).----.)))))))))..))))..).)).)))))))............ ( -33.60) >DroVir_CAF1 36331 102 - 1 CUUAGCUAUGCCCACUUGUCUUGCUGAAGCAGCACCAGGCUG----CCGCCGGCAACUGUCUAACCCAUUUAGCUGAAGACGACUGCGCAUAGCCUGGGCCAGUGC ((..(((((((.((.((((((((((((((((((.....))))----)....(((....))).......))))))..))))))).)).)))))))..))........ ( -36.80) >DroSec_CAF1 21524 96 - 1 CUCAGCUAUGCCCACUUGUCUUGCUGAAUG--U-CAAAGUUUUGAGAGGAAGAAAACU-------GCAUUUAGCUGAAGAUGACUGCGCAUAGUCACGCCCCCAAC ....(((((((.((.(..((((((((((((--(-...((((((.........))))))-------)))))))))..))))..).)).)))))))............ ( -28.40) >DroSim_CAF1 21371 96 - 1 CUCAGCUAUGCCCACUUGUCUUGCUGAAUG--U-CAAAGUUUUGAGAGAAAGAAAACU-------GCAUUUAGCUGAAGAUGACUGCGCAUAGCCACGCCCCCAGC ....(((((((.((.(..((((((((((((--(-...((((((.........))))))-------)))))))))..))))..).)).)))))))............ ( -30.40) >DroEre_CAF1 21374 95 - 1 CUUAGCUAUGCCCACUUGUCUUGCUGAAUG--U-CAAAGUUUUGAGCGGAAGA-AACU-------ACAUUUGGCUGAAGAUGACUGCGCAUAGCCACGCCCUCAGC ....(((((((.((.(..((((((..((((--(-...((((((........))-))))-------)))))..))..))))..).)).)))))))............ ( -30.50) >DroYak_CAF1 21297 96 - 1 CUUAGCUAUGCCCACUUGUCUUGCUGAAUG--U-CAAAGUUUUGCAAGGAAGAAAACU-------ACAUUUAGCUUAAGAUGACUGCGCACAGCCACGCCCCCAUC ....(((.(((.((.(..((((((((((((--(-...((((((.(......)))))))-------)))))))))..))))..).)).))).)))............ ( -26.40) >consensus CUCAGCUAUGCCCACUUGUCUUGCUGAAUG__U_CAAAGUUUUGAGAGGAAGAAAACU_______GCAUUUAGCUGAAGAUGACUGCGCAUAGCCACGCCCCCAGC ....(((((((.((.(((((((((((((((.......((((((.........))))))........))))))))..))))))).)).)))))))............ (-26.18 = -26.41 + 0.23)

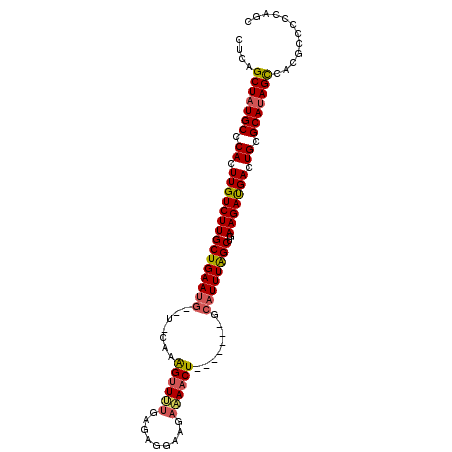
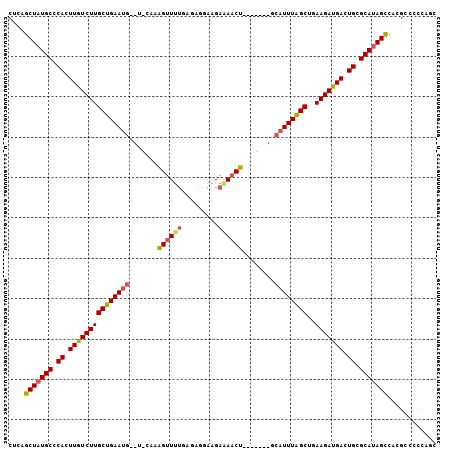
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:04 2006