
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,358,889 – 20,358,992 |
| Length | 103 |
| Max. P | 0.875118 |

| Location | 20,358,889 – 20,358,992 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -25.75 |
| Energy contribution | -25.03 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
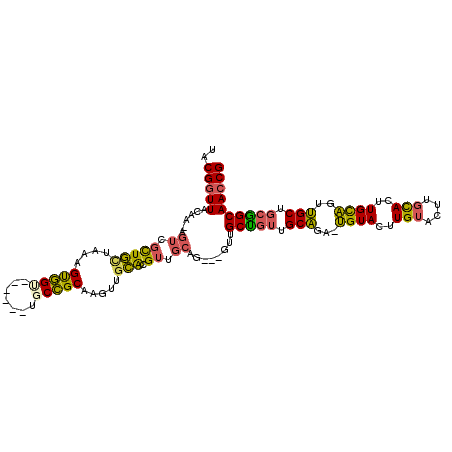
>2L_DroMel_CAF1 20358889 103 + 22407834 UACGGUUACAA-AGUUGCUGCUAAAGUGGU------UGCCGCAAGUUGCACGUUGCAG---GUUGCUGUUGCAGA-UGUACUUGUACUUGCACUUGCAGUUGCUGCGGCAACCG ..((((.((..-.)).)))).......(((------(((((((.((((((.((.((((---((.((...(((...-.)))...)))))))))).))))...)))))))))))). ( -37.90) >DroSec_CAF1 15765 103 + 1 UACGGUUACAG-AGUUGCUGUUAAAGUGGC------UGCCGCAAGUUGCACGUUGCAG---GUUGCUGUUGCAGA-UGUACUUGUACUUGCACUUGCAGUUGCUGCGGCAACCG ..........(-.((((((((...((..((------(((.((((((.(((.((.(((.---.((((....)))).-))))).)))))))))....)))))..))))))))))). ( -40.60) >DroSim_CAF1 14844 103 + 1 UACGGUUACAA-AGUUGCUGCUAAAGCGGC------UGCCGCAAGUUGCACGUUGCAG---GUUGCUGUUGCAGA-UGUACUUGUACUUGCACUUGCAGUUGCUGCGGCAACCG ...........-.((((((((...((((((------(((.((((((.(((.((.(((.---.((((....)))).-))))).)))))))))....))))))))))))))))).. ( -43.00) >DroEre_CAF1 16395 104 + 1 UACGGUUGCAACAGUCGCUGCCAAAGUGGU------UUCCGCAUGUGGCACGUUGCAG---GCUGCCGUUGCAGA-UGUACUUGUACUUGCACUUGCAGUUGCUGCGGCAACCG ..(((((((..((((.(((((((..((((.------..))))...)))))...(((((---(.(((...(((((.-.....)))))...))))))))))).))))..))))))) ( -43.70) >DroYak_CAF1 15663 104 + 1 UACGGUUACAAAAGUCGCUGCCAAAGUGGU------UUCCGCAUGUGGCACGUUGCAG---GUUGCUGUUGCAGA-UGUACUUGUACUUGCACUUGCAGUUGCUGCAGCAACCG .............((.(((((((..((((.------..))))...))))).)).)).(---((((((((.((((.-((((..(((....)))..)))).)))).))))))))). ( -40.70) >DroPer_CAF1 15819 109 + 1 UACGGUUACAGUAGUCGUUACUAAAGUUGUGCCAGUGGCAGCUAGUUCUGGGGUUGAGGCAGUGGCUGUUGCUCGUAGUACUUG---UUGUACUUGUUGUGGCA--AGCAAGCG .(((..((...))..)))(((..((((..(((.((..(((((((.(.((........)).).)))))))..)).)))..)))).---..)))((((((......--)))))).. ( -30.40) >consensus UACGGUUACAA_AGUCGCUGCUAAAGUGGU______UGCCGCAAGUUGCACGUUGCAG___GUUGCUGUUGCAGA_UGUACUUGUACUUGCACUUGCAGUUGCUGCGGCAACCG ..(((((......((.(((((....(((((.......))))).....))).)).))........(((((.(((...((((..(((....)))..))))..))).)))))))))) (-25.75 = -25.03 + -0.71)

| Location | 20,358,889 – 20,358,992 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -19.80 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20358889 103 - 22407834 CGGUUGCCGCAGCAACUGCAAGUGCAAGUACAAGUACA-UCUGCAACAGCAAC---CUGCAACGUGCAACUUGCGGCA------ACCACUUUAGCAGCAACU-UUGUAACCGUA .(((((((((((....((((..((((.(((....))).-..(((....)))..---.))))...))))..))))))))------)))......((((.....-))))....... ( -34.20) >DroSec_CAF1 15765 103 - 1 CGGUUGCCGCAGCAACUGCAAGUGCAAGUACAAGUACA-UCUGCAACAGCAAC---CUGCAACGUGCAACUUGCGGCA------GCCACUUUAACAGCAACU-CUGUAACCGUA .(((((((((((....((((..((((.(((....))).-..(((....)))..---.))))...))))..))))))))------)))......((((.....-))))....... ( -35.70) >DroSim_CAF1 14844 103 - 1 CGGUUGCCGCAGCAACUGCAAGUGCAAGUACAAGUACA-UCUGCAACAGCAAC---CUGCAACGUGCAACUUGCGGCA------GCCGCUUUAGCAGCAACU-UUGUAACCGUA .(((((((((((....((((..((((.(((....))).-..(((....)))..---.))))...))))..))))))))------)))(((.....)))....-........... ( -35.90) >DroEre_CAF1 16395 104 - 1 CGGUUGCCGCAGCAACUGCAAGUGCAAGUACAAGUACA-UCUGCAACGGCAGC---CUGCAACGUGCCACAUGCGGAA------ACCACUUUGGCAGCGACUGUUGCAACCGUA (((((((.((((((.((((...((((.(((....))).-..))))...)))).---.)))..(((((((..((.(...------.)))...))))).))..))).))))))).. ( -40.80) >DroYak_CAF1 15663 104 - 1 CGGUUGCUGCAGCAACUGCAAGUGCAAGUACAAGUACA-UCUGCAACAGCAAC---CUGCAACGUGCCACAUGCGGAA------ACCACUUUGGCAGCGACUUUUGUAACCGUA ((((((((((((....(((...((((.(((....))).-..))))...)))..---))))).(((((((..((.(...------.)))...))))).))......))))))).. ( -34.40) >DroPer_CAF1 15819 109 - 1 CGCUUGCU--UGCCACAACAAGUACAA---CAAGUACUACGAGCAACAGCCACUGCCUCAACCCCAGAACUAGCUGCCACUGGCACAACUUUAGUAACGACUACUGUAACCGUA .(((((((--((........(((((..---...))))).))))))..)))...((((.......(((......))).....))))..((..((((....))))..))....... ( -19.10) >consensus CGGUUGCCGCAGCAACUGCAAGUGCAAGUACAAGUACA_UCUGCAACAGCAAC___CUGCAACGUGCAACUUGCGGCA______ACCACUUUAGCAGCAACU_UUGUAACCGUA ((((((((((((((..((((.((((........))))....(((....)))......))))...)))....))))))................((((......))))))))).. (-19.80 = -21.08 + 1.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:01 2006