
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,357,298 – 20,357,466 |
| Length | 168 |
| Max. P | 0.703446 |

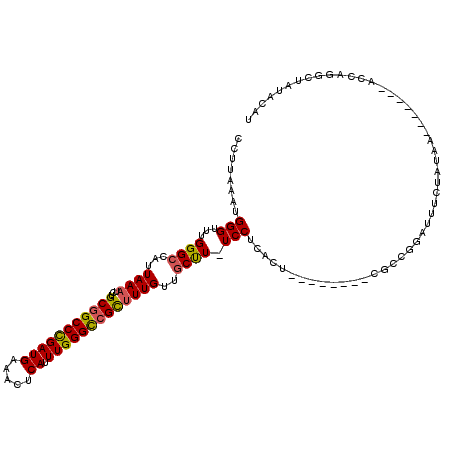
| Location | 20,357,298 – 20,357,405 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.13 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20357298 107 - 22407834 CCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCUUUGUUGCUU-UCCU------------CGUCGGAUUUCUAUAAACCAUAAACCAGGCUAUACGU ((......))((...((((.........(((((((((((.....)).)))))))))(((((.....-(((.------------....))).....)))))..........))))..)).. ( -31.50) >DroSec_CAF1 14176 104 - 1 CCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGUUUUGUUGCUU-UCCUCACU--------CGCCGGAUUUCUAUAA-------ACCAGCCUAUACAU ......(((((((.((............(((((((((((.....)).)))))))))(((((.....-(((.....--------....))).....))))-------)))))))))).... ( -29.10) >DroSim_CAF1 13220 104 - 1 CCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCUUUGCUGCUU-UCCUCACU--------CGCCGGAUUUCUAUAA-------ACCAGGCUAUACAU ........((((..(((...........(((((((((((.....)).)))))))))..........-)))..)))--------)(((((.((.....))-------.)).)))....... ( -30.10) >DroEre_CAF1 14941 104 - 1 CCUUAAAUGGGUUCGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCCUUGCUGCUU-UCCUCACU--------CGUUGGAUUUCUGUAA-------GCCAGGCUAUACAU .((((((((((((((((((...........)))))))....)))))))))))..((((.(((((..-(((.....--------....))).....)).)-------)).))))....... ( -36.20) >DroYak_CAF1 14056 104 - 1 UCUUAAAUGGGUUCGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGUCGCUUUGUUGCUU-UCCUCACU--------CGUUGGAUUUCUAUAC-------AGCAGGCUAUACAU ...((((((((((((((((...........)))))))....)))))))))(((((((.(((.....-(((.....--------....))).....))).-------))).))))...... ( -31.00) >DroAna_CAF1 14396 104 - 1 CCUUAAAUGGGUUUGGGCCAUUAAAUCUGCUGCCUGAUGAAACUCAUUUGGGCUGCUUUGUUACCUUUCCGCCCUGUGGACUCCCCCCGAUUUCUUGAA-------ACU---------AU .......((((...(((...........((.(((..(((.....)).)..))).))...........(((((...)))))..)))))))..........-------...---------.. ( -22.30) >consensus CCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCUUUGUUGCUU_UCCUCACU________CGCCGGAUUUCUAUAA_______ACCAGGCUAUACAU ........(((...((((...((((...(((((((((((.....)).)))))))))))))..)))).))).................................................. (-21.19 = -21.13 + -0.05)


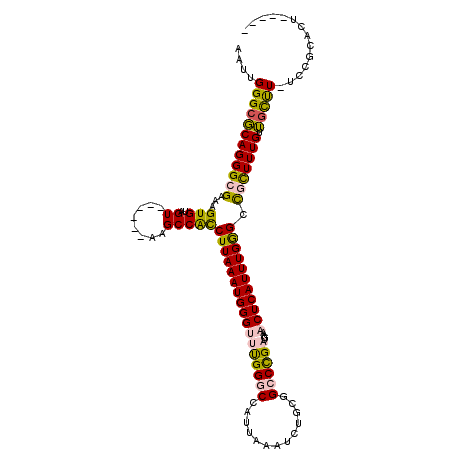
| Location | 20,357,335 – 20,357,437 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -23.40 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20357335 102 - 22407834 AAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCUUUGUUGCUU-UCCU--------- ....((((((((((((...(((....------....)))((((((((((((((((((...........)))))))....))))))))))).))))))).)))))-....--------- ( -35.90) >DroPse_CAF1 14458 111 - 1 AAUUGGGCUCAGGGCGAAAGUGUAGUACGAGUAUGCCGCCUUAAAUGGGUCUGGGCCAUUAAAUUGUUUGGCUGAUGAAUCUCAUUUGAGAC-UUUUGUAGUUU-UUCGGACU----- .....(((((((((((.(..((.....))..).))))((((.....)))))))))))........(((((((((..((.((((....)))).-))...))))..-..))))).----- ( -30.40) >DroSec_CAF1 14206 106 - 1 AAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGUUUUGUUGCUU-UCCUCACU----- ....((((((((((((...(((....------....)))((((((((((((((((((...........)))))))....))))))))))).))))))).)))))-........----- ( -33.40) >DroYak_CAF1 14086 106 - 1 AAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCAUCUUAAAUGGGUUCGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGUCGCUUUGUUGCUU-UCCUCACU----- ...........(((.(((((((....------((((...((((((((((((((((((...........)))))))....)))))))))))..))))...)))))-)))))...----- ( -38.40) >DroAna_CAF1 14420 112 - 1 AAUUGGGCGCAGGGCGAAAGUGUAGU------GUGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCUGCCUGAUGAAACUCAUUUGGGCUGCUUUGUUACCUUUCCGCCCUGUGGA .......(((((((((((((.(((((------(.(((((((.....))))...)))))).......((.(((..(((.....)).)..))).)).....))))))).))))))))).. ( -42.40) >DroPer_CAF1 14389 111 - 1 AAUUGGGCUCAGGGCGAAAGUGUAGUACGAGUAUGCCGCCUUAAAUGGGUCUGGGCCAUUAAAUUGUUUGGCUGAUGAAUCUCAUUUGGGAC-UUUUGUAGUUU-UUCGGACU----- .....(((((((((((.(..((.....))..).))))((((.....)))))))))))........(((((((((..((.((((....)))).-))...))))..-..))))).----- ( -29.50) >consensus AAUUGGGCGCAGGGCGAAAGUGUAGU______AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGAAACUCAUUUGGGCCGCUUUGUUGCUU_UCCGCACU_____ ....((((((((((((...(((..((........)))))(((((((((((((((((.............))))))....))))))))))).))))))).))))).............. (-23.40 = -22.82 + -0.58)

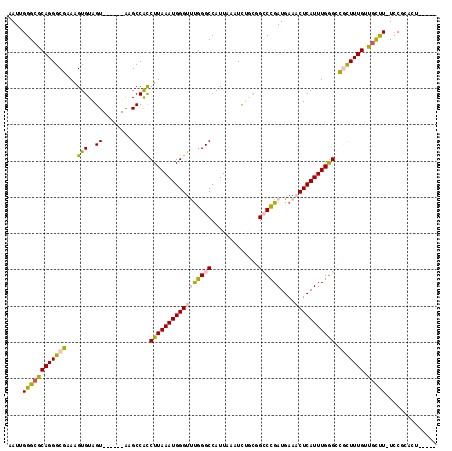
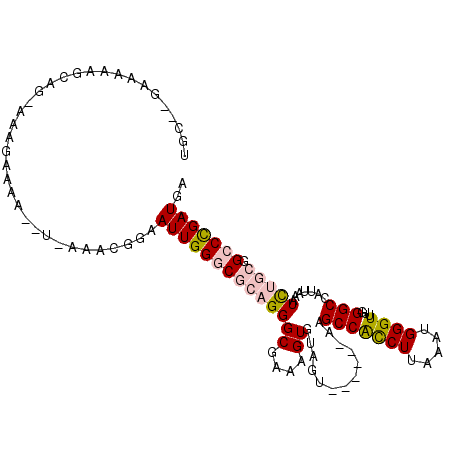
| Location | 20,357,365 – 20,357,466 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20357365 101 - 22407834 UGC--GAAAAAGCAGCUAAGAAAA--U-AAACGGAAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGA (((--......)))..........--.-.......((((((((((((((....)).....------..(((((((.....))))...))).......))))).))))))).. ( -30.10) >DroPse_CAF1 14491 107 - 1 UGCUCGCAAAAGU---AAAGAAAA--CAAAACGGAAUUGGGCUCAGGGCGAAAGUGUAGUACGAGUAUGCCGCCUUAAAUGGGUCUGGGCCAUUAAAUUGUUUGGCUGAUGA ......((..(((---.....(((--(((....((....(((((((((((.(..((.....))..).))))((((.....))))))))))).))...)))))).)))..)). ( -25.30) >DroSec_CAF1 14240 101 - 1 UGC--GAAAAAGCAGGUAACAAAA--U-AAACGGAAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGA (((--......)))..........--.-.......((((((((((((((....)).....------..(((((((.....))))...))).......))))).))))))).. ( -30.10) >DroYak_CAF1 14120 101 - 1 GGC--AAAAAAGCUGCUAAGAAAA--U-AAACGGAAUUGGGCGCAGGGCGAAAGUGUAGU------AAGCCAUCUUAAAUGGGUUCGGGCCAUUAAAUCUGCGGCCCGAUGA (((--(.......)))).......--.-.......((((((((((((((....)).....------..(((((((.....))))...))).......))))).))))))).. ( -27.30) >DroAna_CAF1 14460 102 - 1 UGC--GAAAAAA-AGUAAAGAAACCUA-AAACGGAAUUGGGCGCAGGGCGAAAGUGUAGU------GUGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCUGCCUGAUGA (((--.......-.)))......((..-....)).((..((((((((((....)).((((------(.(((((((.....))))...))))))))..))))).)))..)).. ( -30.70) >DroPer_CAF1 14422 107 - 1 UGCUGGCAAAAGU---AAAGAAAA--CAAAACGGAAUUGGGCUCAGGGCGAAAGUGUAGUACGAGUAUGCCGCCUUAAAUGGGUCUGGGCCAUUAAAUUGUUUGGCUGAUGA .((..((((....---........--(((.......)))(((((((((((.(..((.....))..).))))((((.....)))))))))))......))).)..))...... ( -26.50) >consensus UGC__GAAAAAGCAG_AAAGAAAA__U_AAACGGAAUUGGGCGCAGGGCGAAAGUGUAGU______AAGCCACCUUAAAUGGGUUUGGGCCAUUAAAUCUGCGGCCCGAUGA ...................................((((((((((((((....)).............(((((((.....))))...))).......))))).))))))).. (-19.22 = -19.72 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:00 2006