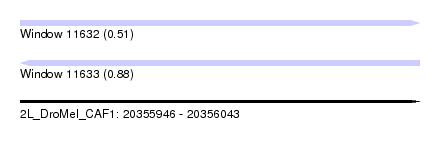
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,355,946 – 20,356,043 |
| Length | 97 |
| Max. P | 0.878354 |

| Location | 20,355,946 – 20,356,043 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.21 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -7.12 |
| Energy contribution | -8.69 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.23 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
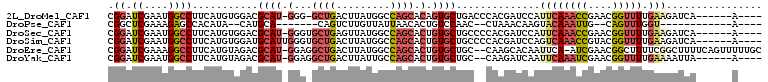
>2L_DroMel_CAF1 20355946 97 + 22407834 ----U------UGAUCUUCAAAACCGUUCGGUUUGAAUGGAUCGUGGGUCAGCACUGUGCUGGCCAUAAGUCAGC-CCC-AUGCGUCCACAUGAAGGCCAUUCGAUCCG ----.------.((((....(((((....)))))((((((.(((((((...(((..(.((((((.....))))))-.).-.)))..)).)))))...)))))))))).. ( -36.80) >DroPse_CAF1 13336 80 + 1 ----U------------ACCAAACUG--CAAUUUGUACUUGUUUAG--GUUGGACAGUGUUAAUAACAAGACUG-------UGCAUG--UAUGUGGCUCUUUCGAGCCG ----.------------(((((((((--(.....)))...)))).)--))((.(((((.((......)).))))-------).))..--.....(((((....))))). ( -20.50) >DroSec_CAF1 12868 98 + 1 ----U------UGAUCUUCAAAACCGUUCGGUUUGAAUGGAUCGUGGGGCAGCACAGUGCUGGCCAUAACUCAGCACCC-AUGCGUCCACAUGAAGGCCAUUCGAUCCG ----.------.((((....(((((....)))))((((((.(((((((((.(((..(((((((.......)))))))..-.))))))).)))))...)))))))))).. ( -39.90) >DroSim_CAF1 11916 99 + 1 ----U------UGAUCUUCAAAACCGUACGGUUUGACUGGAUCGUGGGGCAGCACAGUGCUGGCCAUAAGUCAGCACCCAAUGCAUCCACAUGAAGGCCAUUCGAUCCG ----.------......(((((.((....)))))))..(((((((((((((.....((((((((.....))))))))....))).)))))..(((.....)))))))). ( -37.10) >DroEre_CAF1 13463 105 + 1 GCAAAAACUGAAAAGCCGAAAAGCCGUUCGAU-UGAAUUGUGCUUG--GCAGCACAGUGCUGGCCAUAAGUCAGCCUCC-AUGCGUCUACAUGAAGGCCUUUCGAUCCG .......((....)).(((((.(((.......-...((((((((..--..))))))))((((((.....))))))...(-(((......))))..))).)))))..... ( -29.90) >DroYak_CAF1 12860 96 + 1 ----U------UAAUUUUCAAAACCGUUCGAUUUGAAUUGAUCUUG--GCAGCACAGUGCUGGCAAUAAGUCAGCCUCC-AUGCGUCUACAUGAAGGCCAUUCGAUCCG ----.------.................((..((((((((((.(((--.((((.....)))).)))...))))((((.(-(((......)))).)))).))))))..)) ( -25.10) >consensus ____U______UGAUCUUCAAAACCGUUCGAUUUGAAUGGAUCGUG__GCAGCACAGUGCUGGCCAUAAGUCAGC_CCC_AUGCGUCCACAUGAAGGCCAUUCGAUCCG ...............(((((.........((((......)))).....(((.......((((((.....))))))......))).......)))))............. ( -7.12 = -8.69 + 1.56)


| Location | 20,355,946 – 20,356,043 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.21 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -7.79 |
| Energy contribution | -8.85 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
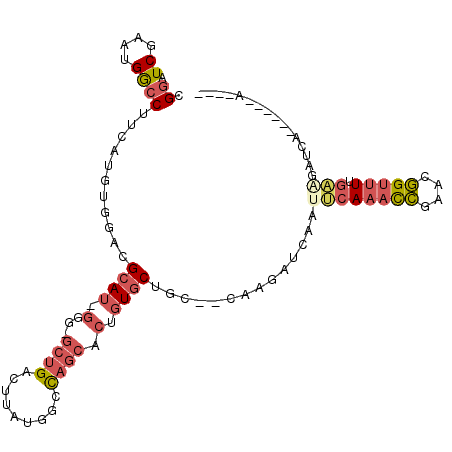
>2L_DroMel_CAF1 20355946 97 - 22407834 CGGAUCGAAUGGCCUUCAUGUGGACGCAU-GGG-GCUGACUUAUGGCCAGCACAGUGCUGACCCACGAUCCAUUCAAACCGAACGGUUUUGAAGAUCA------A---- .((((((...(((((.(((((....))))-)))-))).......((.((((.....)))).))..)))))).((((((((....)).)))))).....------.---- ( -36.40) >DroPse_CAF1 13336 80 - 1 CGGCUCGAAAGAGCCACAUA--CAUGCA-------CAGUCUUGUUAUUAACACUGUCCAAC--CUAAACAAGUACAAAUUG--CAGUUUGGU------------A---- .(((((....))))).....--..((.(-------((((.(((....))).))))).))((--(.((((..(((.....))--).)))))))------------.---- ( -22.70) >DroSec_CAF1 12868 98 - 1 CGGAUCGAAUGGCCUUCAUGUGGACGCAU-GGGUGCUGAGUUAUGGCCAGCACUGUGCUGCCCCACGAUCCAUUCAAACCGAACGGUUUUGAAGAUCA------A---- ..((((((((((...((.((.((..((((-((.(((((.((....)))))))))))))..)).)).)).))))))(((((....)))))....)))).------.---- ( -37.10) >DroSim_CAF1 11916 99 - 1 CGGAUCGAAUGGCCUUCAUGUGGAUGCAUUGGGUGCUGACUUAUGGCCAGCACUGUGCUGCCCCACGAUCCAGUCAAACCGUACGGUUUUGAAGAUCA------A---- .((((((((.....)))..((((..((((..(((((((.(.....).)))))))))))....)))))))))..(((((((....)).)))))......------.---- ( -33.40) >DroEre_CAF1 13463 105 - 1 CGGAUCGAAAGGCCUUCAUGUAGACGCAU-GGAGGCUGACUUAUGGCCAGCACUGUGCUGC--CAAGCACAAUUCA-AUCGAACGGCUUUUCGGCUUUUCAGUUUUUGC ..(((.(((.(((((((((((....))))-)))))))......((((.(((.....)))))--)).......))).-)))(((.((((....)))).)))......... ( -35.20) >DroYak_CAF1 12860 96 - 1 CGGAUCGAAUGGCCUUCAUGUAGACGCAU-GGAGGCUGACUUAUUGCCAGCACUGUGCUGC--CAAGAUCAAUUCAAAUCGAACGGUUUUGAAAAUUA------A---- ..((((....(((((((((((....))))-)))))))......(((.((((.....)))).--)))))))..((((((((....)).)))))).....------.---- ( -31.80) >consensus CGGAUCGAAUGGCCUUCAUGUGGACGCAU_GGG_GCUGACUUAUGGCCAGCACUGUGCUGC__CAAGAUCAAUUCAAACCGAACGGUUUUGAAGAUCA______A____ .((.((....))))...........((((.(...((((.........)))).).))))..............((((((((....))))).)))................ ( -7.79 = -8.85 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:57 2006