
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,350,744 – 20,350,836 |
| Length | 92 |
| Max. P | 0.999899 |

| Location | 20,350,744 – 20,350,836 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.48 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.48 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20350744 92 + 22407834 CCUUGAUGUGUCUGCG---GUGAAAUUCGGCCAA-CUGCCU-GAGGUU--------------AUAUUCAAUGCA--UAUCAGGCGGCCAAGCGAAUUUCACGGCACCUAGA-------CU .........((((((.---((((((((((.....-((((((-((.((.--------------.........)).--..)))))))).....)))))))))).))....)))-------). ( -33.80) >DroSec_CAF1 7755 92 + 1 CCUUGAUGUGUCUGCG---GUGAAAUUCGGCCAA-CUGCCU-GAGGUU--------------AUAUUCAAUGCA--UAUCAGGCGGCCAAGCGAAUUUCACUGCACCUAGA-------CU .........(((((((---((((((((((.....-((((((-((.((.--------------.........)).--..)))))))).....)))))))))))))....)))-------). ( -35.90) >DroEre_CAF1 8222 92 + 1 CCUUGAUGUGUCUGCG---GUGAAAUUCGUCCAA-CUGCCU-GAAGUG--------------ACAUUCAAUGGA--UUUCAGGCGGCCAAGCUAAUUUCACGGCACCUAGA-------CU .........((((((.---((((((((.((....-((((((-(((((.--------------.(((...))).)--))))))))))....)).)))))))).))....)))-------). ( -32.60) >DroYak_CAF1 7663 84 + 1 CCUUGAUGUGUCUGCC---GUGAAAUUCGCCCAA-CUUCCU-GCAGUU--------------G--------GCA--GUUCAGGCAGCCAACCGAAUUUCACGGCACCUAGA-------CU .........(((((((---(((((((((((....-......-)).(((--------------(--------((.--((....)).)))))).))))))))))))....)))-------). ( -32.20) >DroAna_CAF1 7709 120 + 1 CCUUGAUGUGACUCGGCAUGUGGAAGUGGGCCAGGCUGCCCUUUUGUUAAAAAAAAAGACAAAUAUUAAGUAUAAUUUCCGGGCAGCAUGGCCAACUCCAUUGCCUUUAGACCUUAGACU ..............((((.(((((.((.(((((.(((((((.((((((.........))))))(((.....)))......))))))).))))).)))))))))))............... ( -41.20) >consensus CCUUGAUGUGUCUGCG___GUGAAAUUCGGCCAA_CUGCCU_GAAGUU______________AUAUUCAAUGCA__UUUCAGGCGGCCAAGCGAAUUUCACGGCACCUAGA_______CU .......(((((.......((((((((((......((((((.......................................)))))).....))))))))))))))).............. (-17.02 = -17.18 + 0.16)

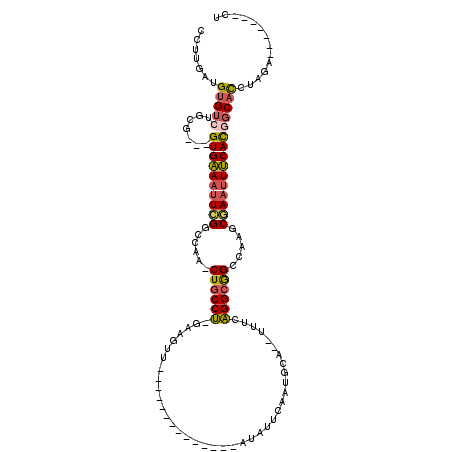
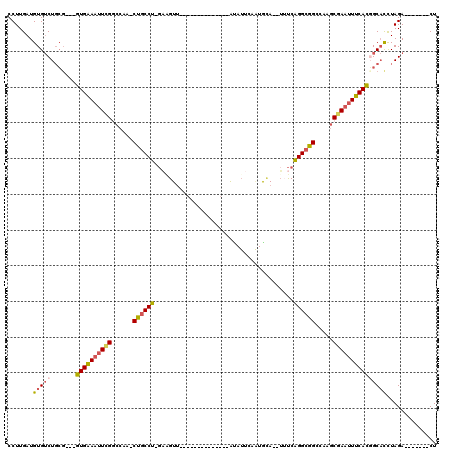
| Location | 20,350,744 – 20,350,836 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.48 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -17.31 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.49 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20350744 92 - 22407834 AG-------UCUAGGUGCCGUGAAAUUCGCUUGGCCGCCUGAUA--UGCAUUGAAUAU--------------AACCUC-AGGCAG-UUGGCCGAAUUUCAC---CGCAGACACAUCAAGG .(-------(((....((.((((((((((.(..((.((((((((--((.......)))--------------)...))-)))).)-)..).))))))))))---.))))))......... ( -33.40) >DroSec_CAF1 7755 92 - 1 AG-------UCUAGGUGCAGUGAAAUUCGCUUGGCCGCCUGAUA--UGCAUUGAAUAU--------------AACCUC-AGGCAG-UUGGCCGAAUUUCAC---CGCAGACACAUCAAGG .(-------(((....((.((((((((((.(..((.((((((((--((.......)))--------------)...))-)))).)-)..).))))))))))---.))))))......... ( -33.40) >DroEre_CAF1 8222 92 - 1 AG-------UCUAGGUGCCGUGAAAUUAGCUUGGCCGCCUGAAA--UCCAUUGAAUGU--------------CACUUC-AGGCAG-UUGGACGAAUUUCAC---CGCAGACACAUCAAGG .(-------(((....((.((((((((.(.(..((.(((((((.--..(((...))).--------------...)))-)))).)-)..).).))))))))---.))))))......... ( -28.60) >DroYak_CAF1 7663 84 - 1 AG-------UCUAGGUGCCGUGAAAUUCGGUUGGCUGCCUGAAC--UGC--------C--------------AACUGC-AGGAAG-UUGGGCGAAUUUCAC---GGCAGACACAUCAAGG .(-------(((....(((((((((((((.(..(((.((((...--...--------.--------------.....)-))).))-)..).))))))))))---)))))))......... ( -38.40) >DroAna_CAF1 7709 120 - 1 AGUCUAAGGUCUAAAGGCAAUGGAGUUGGCCAUGCUGCCCGGAAAUUAUACUUAAUAUUUGUCUUUUUUUUUAACAAAAGGGCAGCCUGGCCCACUUCCACAUGCCGAGUCACAUCAAGG ........(.((...((((.((((((.(((((.(((((((....((((....)))).(((((...........))))).))))))).))))).)).))))..)))).)).)......... ( -41.20) >consensus AG_______UCUAGGUGCCGUGAAAUUCGCUUGGCCGCCUGAAA__UGCAUUGAAUAU______________AACCUC_AGGCAG_UUGGCCGAAUUUCAC___CGCAGACACAUCAAGG ..............(((..((((((((((.(..((.((((.......................................)))).).)..).))))))))))...)))............. (-17.31 = -16.83 + -0.48)

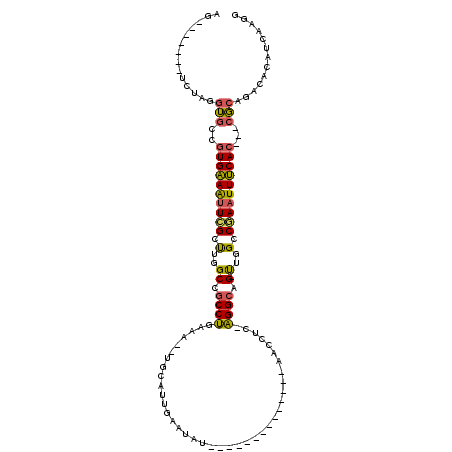
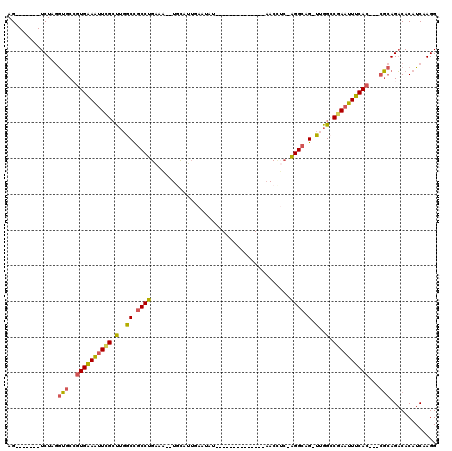
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:54 2006