
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,342,203 – 20,342,357 |
| Length | 154 |
| Max. P | 0.983376 |

| Location | 20,342,203 – 20,342,318 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -18.20 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20342203 115 - 22407834 AAGUAAAAUCGCA---AUAUUUUCGAAUGCCUCGAAAAUUAUAUUACGACAUUGUUACUCACAAA-AACCCCCAAUAUUCCAAAGCUUAAUUGAAAUGGUCUCUUGUAUGAGUGGCACU- ........(((.(---((((((((((.....))))))...))))).)))...((((((((((((.-.(((..((((.............))))....)))...))))..))))))))..- ( -19.62) >DroSec_CAF1 89191 117 - 1 AAGCAAAAUCGCAA--AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA-AAACCUCAAUAUUCCAAAGAUAAAUCCAACUGGUCUCUCGUAUGAGUGACACCU .(((((..((((((--..((((((((.....))))))))....))))))..))))).........-................................(((.(((....))).))).... ( -24.10) >DroSim_CAF1 85178 117 - 1 AAGCAAAAUCGCAA--AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA-AAACAUUAAUAUUCCAAAGCUAAAUCGAAAUGGUCUCAUGUAUGAGUGACACCU .(((((..((((((--..((((((((.....))))))))....))))))..))))).........-...............................(((.((((......)))).))). ( -23.80) >DroEre_CAF1 71872 117 - 1 AAGCAAAAUUGCAAAAAUAUUUUCGAAAAACUCGAGAGUUAUAUUGCGACAUUGCUACCCACAAUCAAACCUAAAUAUUCCAAAGCAAAAUCCAAA---UCUUGUGUAUGAGUGACUCCU .(((((..((((((....((((((((.....))))))))....))))))..)))))...................(((((....((((........---..))))....)))))...... ( -22.20) >consensus AAGCAAAAUCGCAA__AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA_AAACCUCAAUAUUCCAAAGCUAAAUCCAAAUGGUCUCUUGUAUGAGUGACACCU .(((((..((((((....((((((((.....))))))))....))))))..)))))..........................................(((.(((....))).))).... (-18.20 = -17.95 + -0.25)



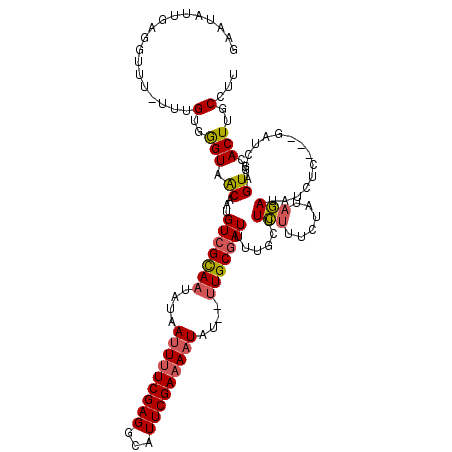
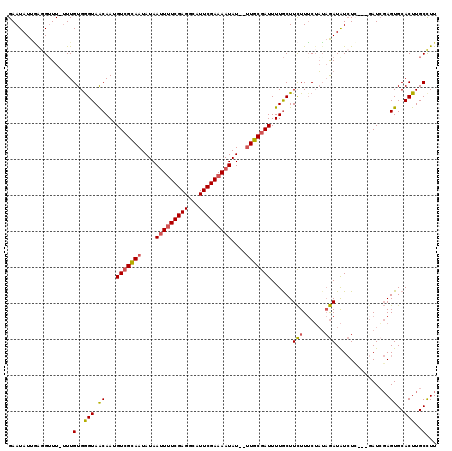
| Location | 20,342,242 – 20,342,357 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20342242 115 + 22407834 GAAUAUUGGGGGUU-UUUGUGAGUAACAAUGUCGUAAUAUAAUUUUCGAGGCAUUCGAAAAUAU---UGCGAUUUUACUUCUUUCUAUAGAUAUCUGAUGUAUAGAGUGCACUUGCCUU ........(((((.-..(((((((((....((((((((((...(((((((...)))))))))))---)))))).))))))..(((((((.((.....)).))))))).)))...))))) ( -28.50) >DroSec_CAF1 89231 113 + 1 GAAUAUUGAGGUUU-UUUGUGGGUAACAAUGUCGCAAUAUAAUUUUCGAGGCAUUCGAAAAUAU--UUGCGAUUUUGCUUCUUUCUAUAGAUAUCUC---GAUCGAGUGCACUUGCCUU ....((((((((..-((((((((...(((.(((((((....(((((((((...)))))))))..--))))))).)))......)))))))).)))))---))).(((.((....))))) ( -31.20) >DroSim_CAF1 85218 113 + 1 GAAUAUUAAUGUUU-UUUGUGGGUAACAAUGUCGCAAUAUAAUUUUCGAGGCAUUCGAAAAUAU--UUGCGAUUUUGCUUCUUUCUAUAGAUAUCUC---GAUCGAGUGCACUUGCCUU ..............-.....((((((....(((((((....(((((((((...)))))))))..--)))))))..(((.(((......)))...(((---....))).))).)))))). ( -23.30) >DroEre_CAF1 71909 115 + 1 GAAUAUUUAGGUUUGAUUGUGGGUAGCAAUGUCGCAAUAUAACUCUCGAGUUUUUCGAAAAUAUUUUUGCAAUUUUGCUUUUUACGUGCAAUGU-UG---GAUCUAGUGCACUUGCUUU ......(((((((..((((((.(((((((.((.((((........(((((...)))))........)))).)).)))))....)).))))..))-..---))))))).((....))... ( -24.09) >consensus GAAUAUUGAGGUUU_UUUGUGGGUAACAAUGUCGCAAUAUAAUUUUCGAGGCAUUCGAAAAUAU__UUGCGAUUUUGCUUCUUUCUAUAGAUAUCUC___GAUCGAGUGCACUUGCCUU ..................(..(((.((...(((((((....(((((((((...)))))))))....)))))))......(((......)))...............))..)))..)... (-17.06 = -17.38 + 0.31)

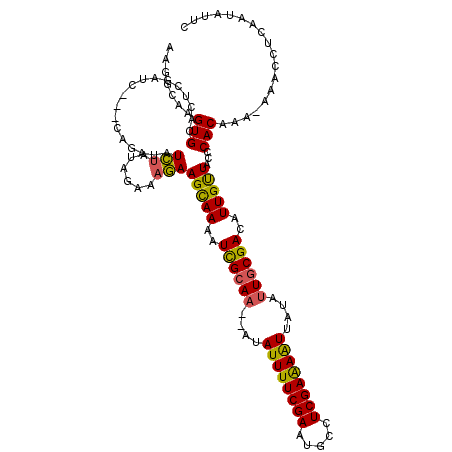
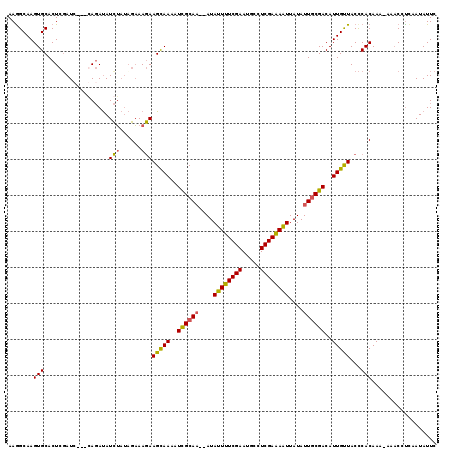
| Location | 20,342,242 – 20,342,357 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20342242 115 - 22407834 AAGGCAAGUGCACUCUAUACAUCAGAUAUCUAUAGAAAGAAGUAAAAUCGCA---AUAUUUUCGAAUGCCUCGAAAAUUAUAUUACGACAUUGUUACUCACAAA-AACCCCCAAUAUUC .......(((...((((((.((.....)).))))))....(((((..(((.(---((((((((((.....))))))...))))).))).....))))))))...-.............. ( -14.70) >DroSec_CAF1 89231 113 - 1 AAGGCAAGUGCACUCGAUC---GAGAUAUCUAUAGAAAGAAGCAAAAUCGCAA--AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA-AAACCUCAAUAUUC .......(((..(((....---)))...............(((((..((((((--..((((((((.....))))))))....))))))..)))))...)))...-.............. ( -22.30) >DroSim_CAF1 85218 113 - 1 AAGGCAAGUGCACUCGAUC---GAGAUAUCUAUAGAAAGAAGCAAAAUCGCAA--AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA-AAACAUUAAUAUUC .......(((..(((....---)))...............(((((..((((((--..((((((((.....))))))))....))))))..)))))...)))...-.............. ( -22.30) >DroEre_CAF1 71909 115 - 1 AAAGCAAGUGCACUAGAUC---CA-ACAUUGCACGUAAAAAGCAAAAUUGCAAAAAUAUUUUCGAAAAACUCGAGAGUUAUAUUGCGACAUUGCUACCCACAAUCAAACCUAAAUAUUC .......(((((.......---..-....)))))......(((((..((((((....((((((((.....))))))))....))))))..)))))........................ ( -23.72) >consensus AAGGCAAGUGCACUCGAUC___CAGAUAUCUAUAGAAAGAAGCAAAAUCGCAA__AUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAAA_AAACCUCAAUAUUC .......(((..................(((......)))(((((..((((((....((((((((.....))))))))....))))))..)))))...))).................. (-17.76 = -17.20 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:51 2006