
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,333,788 – 20,333,888 |
| Length | 100 |
| Max. P | 0.530611 |

| Location | 20,333,788 – 20,333,888 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -21.05 |
| Energy contribution | -20.67 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20333788 100 + 22407834 UCAGUGCAAGUUGUGCCAGCGCACGGUCUGCGCCAAGUGCUAUACCAAGGUAGGUAC----CGGGUUAAUGGUUAAC-CAUAGGGCUGCAUAUUUACUAUUUUC----G ...(((((..((((((....))))))..)))))...((((..((((......))))(----(.(((((.....))))-)...))...)))).............----. ( -29.30) >DroPse_CAF1 48376 107 + 1 UCAGUGCAAGCUCUGCCAGCGCACUGUCUGCGCCAAGUGCUAUACCAAGGUGGGUGGAAUCUGGAACACUAACUGACUCAUUUCAUUGUCUAAUCAC--UUUCUUAUUG .((((((..(((.....)))))))))...((((...))))......((((((..((((...(((((..............)))))...))))..)))--)))....... ( -22.74) >DroSec_CAF1 80774 100 + 1 CCAGUGCAAGCUGUGCCAGCGCACAGUCUGCGCCAAGUGCUAUACCAAGGUGGGUAC----CGGGUUAUUGGUUAAC-CAUAUGGCUGCAUAUUUACACAUUCC----G ...(((((..((((((....))))))..)))))...((((....(((..((((..((----(((....)))))...)-))).)))..)))).............----. ( -34.50) >DroSim_CAF1 76818 100 + 1 CCAGUGCAAGCUGUGCCAGCGCACGGUCUGCGCCAAGUGCUAUACCAAGGUGGGUAC----CGGGUUAUUGGUUAAC-CAUAUGGCUGCAUAUUUACUGUUUCC----G .(((((((..((((((....))))))..)))((((.(((((.(((....))))))))----..(((((.....))))-)...)))).........)))).....----. ( -35.20) >DroEre_CAF1 63429 98 + 1 CCAGUGCAAGCUGUGCCAGCGCACGGUCUGCGCCAAGUGCUAUACCAAGGUGGGUUU----UUGGGUAAUGGUUAAC-CCUAGGCCUGCAUAUUUACAAUUU------G ...(((((..((((((....))))))..)))))(((((...........(..(((((----..((((........))-)).)))))..).........))))------) ( -33.65) >DroYak_CAF1 57089 100 + 1 UCAGUGCAAGCUGUGCCAGCGCACGGUCUGCGCCAAGUGCUAUACCAAGGUGGGUAU----UUGGCUAAUGGUUAUC-CCUAGGCCAACAUAUUUAUAAUUUCU----G .....(((..((((((....))))))..)))((((((((((.(((....))))))))----)))))...(((((...-....))))).................----. ( -34.50) >consensus CCAGUGCAAGCUGUGCCAGCGCACGGUCUGCGCCAAGUGCUAUACCAAGGUGGGUAC____CGGGUUAAUGGUUAAC_CAUAGGGCUGCAUAUUUACAAUUUCC____G ((.(((((..((((((....))))))..)))))...(((((.(((....))))))))......))............................................ (-21.05 = -20.67 + -0.38)

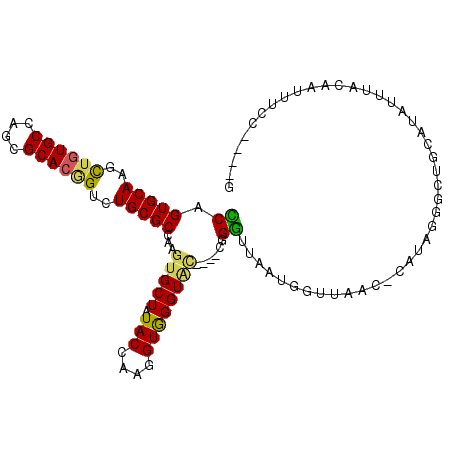
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:45 2006