
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,099,193 – 2,099,290 |
| Length | 97 |
| Max. P | 0.808973 |

| Location | 2,099,193 – 2,099,290 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
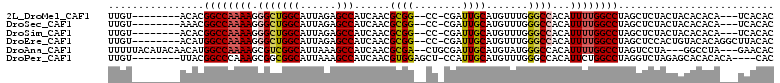
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -22.33 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2099193 97 + 22407834 UUGU--------ACACGGCCAAAAGGGCUGGCAUUAGAGCCAUCAACGCGG--CC-CGAUUGCAUGUUUGGGCCACAUUUUGGCCUAGCUCUACUACACACA---UCACAC ....--------....((((((((.((.((((......))))))...(.((--((-(((........))))))).).)))))))).................---...... ( -31.90) >DroSec_CAF1 116264 97 + 1 UUGU--------AAACGGCCAAAAGGGCUGGCAUUAGAGCCAUCAACGCGG--CC-CGAUUGCAUGUUUGGGCCACAUUUUGGCCUAGCUCUACUACACACA---UCACAC ....--------....((((((((.((.((((......))))))...(.((--((-(((........))))))).).)))))))).................---...... ( -31.90) >DroSim_CAF1 116140 97 + 1 UUGU--------ACACGGCCAAAAGGGCUGGCAUUAGAGCCAUCAACGCGG--CC-CGAUUGCAUGUUUGGGCCACAUUUUGGCCUAGCUCUACUACACACA---UCACAC ....--------....((((((((.((.((((......))))))...(.((--((-(((........))))))).).)))))))).................---...... ( -31.90) >DroEre_CAF1 118895 100 + 1 UUGU--------ACAUGGCCAAAAGGGCUGGCAUUAGAGCCAUCAACGCGG--CC-CGAUUGCAUGUUUGGGCCACAUUUUGGCCUAGCUCCACUGUACACAGGCUUACAC .(((--------(((.((((.....))))((..((((.((((.....(.((--((-(((........))))))).)....))))))))..))..))))))........... ( -35.30) >DroAna_CAF1 120012 103 + 1 UUUUUACAUACAACAUGGCCAAAAGCGUCGGCAUUAAAGCCAUCAACGCGA--CUGCGAUUGCAUGUAUGGGCCACAUUUUGGCCUAGUCCUA---GGCCUA---GAACAC ................((((....((((.(((......)))....))))((--((((....)))....(((((((.....))))))))))...---))))..---...... ( -31.30) >DroPer_CAF1 171449 98 + 1 UUGU--------UUACGGCCCAAAGCGGCGGCAUUAAAGCCAUCAACGUGGAGCU-CCAUUGCAUGUUUGGGCCACAUUCUGGCCUAGGUCUAGAGCACACACA----CAC ....--------....((((((((...(((((......)))......((((....-)))).))...))))))))...(((((((....).))))))........----... ( -31.50) >consensus UUGU________ACACGGCCAAAAGGGCUGGCAUUAGAGCCAUCAACGCGG__CC_CGAUUGCAUGUUUGGGCCACAUUUUGGCCUAGCUCUACUACACACA___UCACAC ................((((((((.(((((((......)))......((((........)))).......))))...)))))))).......................... (-22.33 = -22.55 + 0.22)
| Location | 2,099,193 – 2,099,290 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
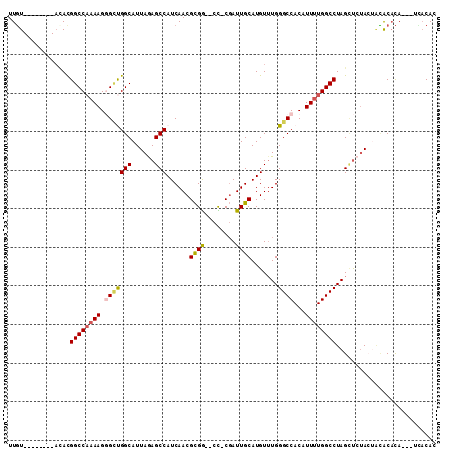
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2099193 97 - 22407834 GUGUGA---UGUGUGUAGUAGAGCUAGGCCAAAAUGUGGCCCAAACAUGCAAUCG-GG--CCGCGUUGAUGGCUCUAAUGCCAGCCCUUUUGGCCGUGU--------ACAA ......---((((..(((.....)).((((((((((((((((............)-))--)))))))..((((......))))......)))))))..)--------))). ( -36.00) >DroSec_CAF1 116264 97 - 1 GUGUGA---UGUGUGUAGUAGAGCUAGGCCAAAAUGUGGCCCAAACAUGCAAUCG-GG--CCGCGUUGAUGGCUCUAAUGCCAGCCCUUUUGGCCGUUU--------ACAA .(((((---......(((.....)))((((((((((((((((............)-))--)))))))..((((......))))......))))))..))--------))). ( -36.30) >DroSim_CAF1 116140 97 - 1 GUGUGA---UGUGUGUAGUAGAGCUAGGCCAAAAUGUGGCCCAAACAUGCAAUCG-GG--CCGCGUUGAUGGCUCUAAUGCCAGCCCUUUUGGCCGUGU--------ACAA ......---((((..(((.....)).((((((((((((((((............)-))--)))))))..((((......))))......)))))))..)--------))). ( -36.00) >DroEre_CAF1 118895 100 - 1 GUGUAAGCCUGUGUACAGUGGAGCUAGGCCAAAAUGUGGCCCAAACAUGCAAUCG-GG--CCGCGUUGAUGGCUCUAAUGCCAGCCCUUUUGGCCAUGU--------ACAA .((((((((..(.....)..).))).((((((((((((((((............)-))--)))))))..((((......))))......))))))...)--------))). ( -38.80) >DroAna_CAF1 120012 103 - 1 GUGUUC---UAGGCC---UAGGACUAGGCCAAAAUGUGGCCCAUACAUGCAAUCGCAG--UCGCGUUGAUGGCUUUAAUGCCGACGCUUUUGGCCAUGUUGUAUGUAAAAA ......---..((((---((....))))))..(((((((((......(((....))).--..(((((...(((......))))))))....)))))))))........... ( -35.50) >DroPer_CAF1 171449 98 - 1 GUG----UGUGUGUGCUCUAGACCUAGGCCAGAAUGUGGCCCAAACAUGCAAUGG-AGCUCCACGUUGAUGGCUUUAAUGCCGCCGCUUUGGGCCGUAA--------ACAA ...----.(((.(.((((((......(((((.....)))))...........)))-))).))))(((.(((((((....((....))...))))))).)--------)).. ( -30.53) >consensus GUGUGA___UGUGUGUAGUAGAGCUAGGCCAAAAUGUGGCCCAAACAUGCAAUCG_GG__CCGCGUUGAUGGCUCUAAUGCCAGCCCUUUUGGCCGUGU________ACAA ..........................(((((((((((((.....................)))))))..((((......))))......))))))................ (-20.92 = -20.45 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:02 2006