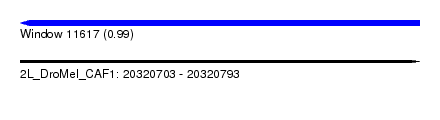
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,320,703 – 20,320,793 |
| Length | 90 |
| Max. P | 0.988021 |

| Location | 20,320,703 – 20,320,793 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -20.04 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
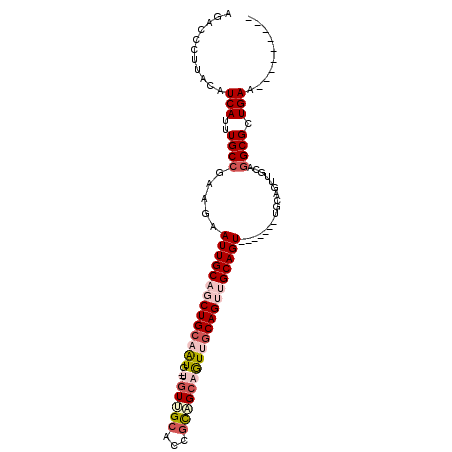
>2L_DroMel_CAF1 20320703 90 - 22407834 AGACCCUUACAUCAUUUGCCGAAGAAUUGCAGCUGCAAUGCCUGUUGCACCGCAGCAGUUGCAGUUGCAGU-------UGCAGUUGCAGGCGUUGAA--------- ...........(((..((((....((((((((((((((((((((((((...)))))))..))).)))))))-------)))))))...)))).))).--------- ( -39.50) >DroSec_CAF1 67746 84 - 1 AGACCCUUACAUCAUUUGCCGAAGAAUUGCAGCUGCAAUGCCUGUUGCACCGCUGCAGUUGCAGUUGCAGU-------------UGCAGGCGUUGAA--------- ...........(((..((((....((((((((((((((...((((.((...)).)))))))))))))))))-------------)...)))).))).--------- ( -32.00) >DroEre_CAF1 50255 87 - 1 AGACCCUUACAUCAUUUGCCGAAGAAUUGCAUCUGCAGU---UGUUGCACCGCAGCAGUCGCAGUUGCAGU-------UGCAGUUGCAGGCGCUGAA--------- ...........(((..((((....(((((((.((((..(---((((((...)))))))..)))).))))))-------)((....)).)))).))).--------- ( -31.40) >DroYak_CAF1 43734 87 - 1 AGACCCUUACAUCAUUUGCCAAAGAAUUGCAUCUGCAGU---UGUUGCACCUCAGCAAUUGCAGUUGCAGU-------UGCAGUUGAAGGCGCUGAA--------- ((.(((((.((....((((.....(((((((.(((((((---(((((.....)))))))))))).))))))-------))))).)))))).)))...--------- ( -31.00) >DroAna_CAF1 32718 91 - 1 GGGCCCUUACAUCAUUUGCCGCAGAAUUGCAGCUGCAAUG----UGGCA--GUUGCAGUUGCAGUGGCAGUGGCACUCUGCAGUUGCAGGCGUUGAA--------- ..(((...........((((((.(.((((((((((((((.----.....--))))))))))))))..).))))))...(((....))))))......--------- ( -37.60) >DroPer_CAF1 33979 96 - 1 -GGCCCUUACAUCAUUUGCAGAAGAAUUGCAGCUGCAGUGGCAGUGGCA--GUGGCAGUGACAGUGGCAGU-------GACAGUUGCAAGCGCUGAGUGGAGGGGA -..(((((...((((...(((.....(((((((((((.((.((.((.((--.......)).)).)).)).)-------).)))))))))...))).))))))))). ( -34.20) >consensus AGACCCUUACAUCAUUUGCCGAAGAAUUGCAGCUGCAAUG__UGUUGCACCGCAGCAGUUGCAGUUGCAGU_______UGCAGUUGCAGGCGCUGAA_________ ...........(((..((((.....((((((((((((((...((((((...)))))))))))))))))))).................)))).))).......... (-20.04 = -21.60 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:42 2006