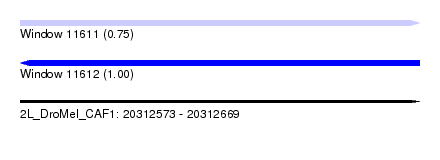
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,312,573 – 20,312,669 |
| Length | 96 |
| Max. P | 0.995956 |

| Location | 20,312,573 – 20,312,669 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.75 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20312573 96 + 22407834 UGGGUGCGCCAC---------CAACCCGAGGGAAUCGAAGUCGGAGUCCAAGUCGGAAAUCGGUGGCUUUGGCGGCUU-UUUCGGGGCCGCCAAAUAGGAUUACUU .(((((.(((((---------(...((((.(((.(((....)))..)))...)))).....))))))(((((((((((-.....)))))))))))......))))) ( -39.90) >DroSec_CAF1 55516 94 + 1 UGGGUGCGCCAC---------CUACCCGAGGGAAUCGAAGUCGGAGUCCAAGUCGGA-AUCGGUGGCUUUGGCGGCUU--UUCGGGGCCGCCAAAUAGGAUAACUU .((((.((((((---------(...((((.(((.(((....)))..)))...)))).-...))))))(((((((((((--....)))))))))))...)...)))) ( -39.80) >DroSim_CAF1 54332 94 + 1 UGGGUGCGCCAC---------CUACCCGAGGGAAUCGAAGUCGGAGUCCAAGUCGGA-AUCGGUGGCUUUGGCGGCUU--GUCGGGGCCGCCAAAUAGGAUUACUU .(((((.(((((---------(...((((.(((.(((....)))..)))...)))).-...))))))((((((((((.--.....))))))))))......))))) ( -40.20) >DroEre_CAF1 41751 83 + 1 UGGGUGCGCCAC---------CUACCCGAGGGAAUCGAAGUUGGA-----------A-AUCGGUGACUUUGGCGGCUU--UUCGGGGCCGCCAAAUAGGAUUACUU ((((((...)))---------))).((((((..(((((.......-----------.-.)))))..))))((((((((--....)))))))).....))....... ( -29.10) >DroYak_CAF1 35503 83 + 1 UGGGUGCGCCAC---------CUACCCGAGGGAAUCGAAGUCGGA-----------A-AUCGGUGGCUUUGGCGGCUU--UUCGGGGCCGCCAAAUAGGAUUACUU .(((((.(((((---------(..((....))..(((....))).-----------.-...))))))(((((((((((--....)))))))))))......))))) ( -34.50) >DroPer_CAF1 26276 90 + 1 CGGGUGCGGCUCCACAGAUCCCUGCCCCGCAGAGCCGACAC---------------A-UCCGACAGAUGUUGAGGCAAGUCUGAGGUCCGCCAAAUAGGAUUACCU .((((((((..((.(((((..((((...)))).(((...((---------------(-((.....)))))...)))..))))).)).))))...........)))) ( -29.40) >consensus UGGGUGCGCCAC_________CUACCCGAGGGAAUCGAAGUCGGA___________A_AUCGGUGGCUUUGGCGGCUU__UUCGGGGCCGCCAAAUAGGAUUACUU .(((((..((...........((((((((.....)))..........((.....)).....))))).((((((((((........))))))))))..))..))))) (-18.47 = -19.75 + 1.28)

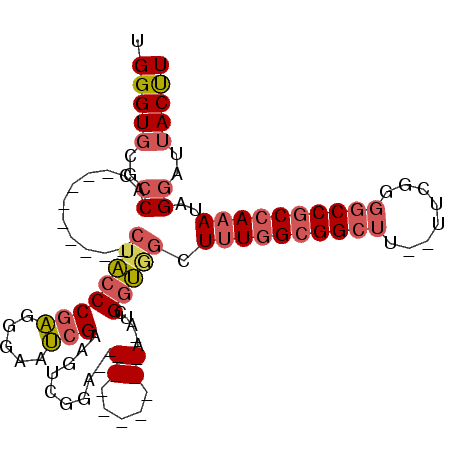
| Location | 20,312,573 – 20,312,669 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20312573 96 - 22407834 AAGUAAUCCUAUUUGGCGGCCCCGAAA-AAGCCGCCAAAGCCACCGAUUUCCGACUUGGACUCCGACUUCGAUUCCCUCGGGUUG---------GUGGCGCACCCA ..((.......(((((((((.......-..)))))))))(((((((((..((((...(((...((....))..))).))))))))---------)))))))..... ( -38.50) >DroSec_CAF1 55516 94 - 1 AAGUUAUCCUAUUUGGCGGCCCCGAA--AAGCCGCCAAAGCCACCGAU-UCCGACUUGGACUCCGACUUCGAUUCCCUCGGGUAG---------GUGGCGCACCCA ...........(((((((((......--..)))))))))((((((...-(((((...(((...((....))..))).)))))..)---------)))))....... ( -38.50) >DroSim_CAF1 54332 94 - 1 AAGUAAUCCUAUUUGGCGGCCCCGAC--AAGCCGCCAAAGCCACCGAU-UCCGACUUGGACUCCGACUUCGAUUCCCUCGGGUAG---------GUGGCGCACCCA ..((.......(((((((((......--..)))))))))((((((...-(((((...(((...((....))..))).)))))..)---------)))))))..... ( -38.80) >DroEre_CAF1 41751 83 - 1 AAGUAAUCCUAUUUGGCGGCCCCGAA--AAGCCGCCAAAGUCACCGAU-U-----------UCCAACUUCGAUUCCCUCGGGUAG---------GUGGCGCACCCA ..((.......(((((((((......--..)))))))))((((((((.-.-----------.......))....((....))..)---------)))))))..... ( -27.20) >DroYak_CAF1 35503 83 - 1 AAGUAAUCCUAUUUGGCGGCCCCGAA--AAGCCGCCAAAGCCACCGAU-U-----------UCCGACUUCGAUUCCCUCGGGUAG---------GUGGCGCACCCA ..((.......(((((((((......--..)))))))))((((((...-.-----------(((((...........)))))..)---------)))))))..... ( -33.50) >DroPer_CAF1 26276 90 - 1 AGGUAAUCCUAUUUGGCGGACCUCAGACUUGCCUCAACAUCUGUCGGA-U---------------GUGUCGGCUCUGCGGGGCAGGGAUCUGUGGAGCCGCACCCG (((....))).....((((.((.((((((((((((.(((((.....))-)---------------))....((...))))))))))..)))).))..))))..... ( -34.00) >consensus AAGUAAUCCUAUUUGGCGGCCCCGAA__AAGCCGCCAAAGCCACCGAU_U___________UCCGACUUCGAUUCCCUCGGGUAG_________GUGGCGCACCCA ..((.......(((((((((..........)))))))))((((((((.....................)))...((....))............)))))))..... (-18.16 = -18.67 + 0.50)
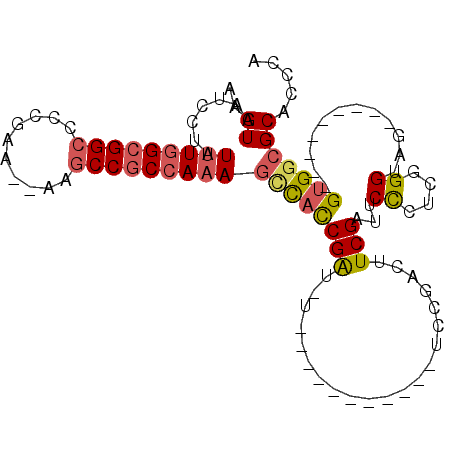
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:37 2006