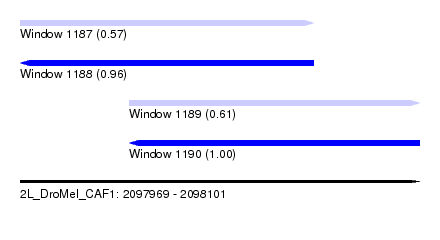
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,097,969 – 2,098,101 |
| Length | 132 |
| Max. P | 0.999053 |

| Location | 2,097,969 – 2,098,066 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.59 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
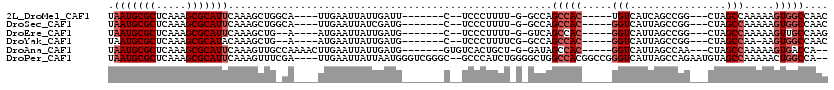
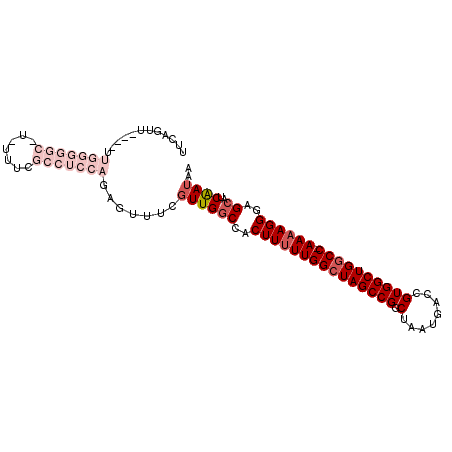
>2L_DroMel_CAF1 2097969 97 + 22407834 UAAUGCGCUCAAAGCGCAUUCAAAGCUGGCA----UUGAAUUAUUGAUU-------C--UCCCUUUU-G-GCCAGCCAC-----UGUCAUCAGCCGG---CUAGCCAAAAAGUGGCCAAC .(((((((.....)))))))......((((.----..(((((...))))-------)--....((((-(-((.((((.(-----((....)))..))---)).)))))))....)))).. ( -34.20) >DroSec_CAF1 115060 97 + 1 UAAUGCGCUCAAAGCGCAUUCAAAGCUGGCA----UUGAAUUAUCGAUG-------C--UCCCUUUU-G-GCCAGCCAC-----GGUCAUUAGCCGG---CUAGCCAAAAAGUGGCCAAC .(((((((.....)))))))....(((((((----((((....))))))-------)--)...((((-(-((.((((..-----(((.....)))))---)).)))))))...))).... ( -37.00) >DroEre_CAF1 117680 95 + 1 UAAUGCGCUCAAAGCGCAUUCAAAGCUG--A----AUGAAUUAUUGAUG-------C--UCCCUUUU-G-GUCAGCCAC-----GGUCAUUAGCCGG---CUAGCCAAAAAGUUGCCAAG .(((((((.....)))))))........--.----.............(-------(--..(.((((-(-(..((((..-----(((.....)))))---))..)))))).)..)).... ( -27.70) >DroYak_CAF1 119818 95 + 1 UAAUGCGCUCAAAGCGCAUACAAAGCUG--A----AUGAAUUAUUGAUG-------C--UCCCUUUUCG-GCCAGCCAC-----GGUCAUUAGCCGG---CUAGCCAA-AAGUGGCCAAC ..((((((.....))))))....((((.--(----(((...)))).).)-------)--)(((((((.(-((.((((..-----(((.....)))))---)).)))))-))).))..... ( -28.30) >DroAna_CAF1 118791 101 + 1 UAAUGCGCUCAAAGCGCAUUCAAAGUUGCCAAAACUUGAAUUAUUGAUG-------GUGUCACUGCU-G-GAUAGCCAC-----GGUCAUUAGCCAA---CUAGCCAAAAAGUGACCA-- ...(((((.....)))))(((((.(((.....)))))))).........-------(.((((((..(-(-(.(((....-----(((.....)))..---))).)))...))))))).-- ( -26.40) >DroPer_CAF1 169797 112 + 1 UAAUGCGCUCAAAGCGCAUUCAAAGUUUCGA----UUGAAUUAUUAAUGGGUCGGGC--GCCCAUCUGGGGCUGGCCACGGCCGGGUCAUUAGCCAGAAUGUAGCCAAAAACUGGCCA-- .(((((((.....)))))))......(((..----..............((((((((--((((.....))))..))).))))).(((.....))).)))....((((.....))))..-- ( -37.50) >consensus UAAUGCGCUCAAAGCGCAUUCAAAGCUG_CA____UUGAAUUAUUGAUG_______C__UCCCUUUU_G_GCCAGCCAC_____GGUCAUUAGCCGG___CUAGCCAAAAAGUGGCCAAC .(((((((.....)))))))......................................................(((((.....(((................))).....))))).... (-13.76 = -14.59 + 0.83)
| Location | 2,097,969 – 2,098,066 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2097969 97 - 22407834 GUUGGCCACUUUUUGGCUAG---CCGGCUGAUGACA-----GUGGCUGGC-C-AAAAGGGA--G-------AAUCAAUAAUUCAA----UGCCAGCUUUGAAUGCGCUUUGAGCGCAUUA ((((((..((((((((((((---(((.(((....))-----)))))))))-)-))))))..--(-------(((.....))))..----.))))))....(((((((.....))))))). ( -47.90) >DroSec_CAF1 115060 97 - 1 GUUGGCCACUUUUUGGCUAG---CCGGCUAAUGACC-----GUGGCUGGC-C-AAAAGGGA--G-------CAUCGAUAAUUCAA----UGCCAGCUUUGAAUGCGCUUUGAGCGCAUUA ((((((..((((((((((((---(((((....).))-----..)))))))-)-))))))..--.-------....((....))..----.))))))....(((((((.....))))))). ( -43.60) >DroEre_CAF1 117680 95 - 1 CUUGGCAACUUUUUGGCUAG---CCGGCUAAUGACC-----GUGGCUGAC-C-AAAAGGGA--G-------CAUCAAUAAUUCAU----U--CAGCUUUGAAUGCGCUUUGAGCGCAUUA ....((..((((((((.(((---(((((....).))-----..))))).)-)-))))))..--)-------).............----.--........(((((((.....))))))). ( -31.40) >DroYak_CAF1 119818 95 - 1 GUUGGCCACUU-UUGGCUAG---CCGGCUAAUGACC-----GUGGCUGGC-CGAAAAGGGA--G-------CAUCAAUAAUUCAU----U--CAGCUUUGUAUGCGCUUUGAGCGCAUUA (.((.((..((-((((((((---(((((....).))-----..)))))))-)))))..)).--.-------)).)..........----.--.........((((((.....)))))).. ( -32.30) >DroAna_CAF1 118791 101 - 1 --UGGUCACUUUUUGGCUAG---UUGGCUAAUGACC-----GUGGCUAUC-C-AGCAGUGACAC-------CAUCAAUAAUUCAAGUUUUGGCAACUUUGAAUGCGCUUUGAGCGCAUUA --..((((((..((((.(((---(((((....).))-----)..)))).)-)-)).))))))..-------........(((((((((.....))).))))))((((.....)))).... ( -27.50) >DroPer_CAF1 169797 112 - 1 --UGGCCAGUUUUUGGCUACAUUCUGGCUAAUGACCCGGCCGUGGCCAGCCCCAGAUGGGC--GCCCGACCCAUUAAUAAUUCAA----UCGAAACUUUGAAUGCGCUUUGAGCGCAUUA --.((((.(((.(((((((.....))))))).)))..))))(.(((..((((.....))))--))))............((((((----........))))))((((.....)))).... ( -37.90) >consensus GUUGGCCACUUUUUGGCUAG___CCGGCUAAUGACC_____GUGGCUGGC_C_AAAAGGGA__G_______CAUCAAUAAUUCAA____UG_CAGCUUUGAAUGCGCUUUGAGCGCAUUA ..((((((.....))))))....(((((((............)))))))...................................................(((((((.....))))))). (-21.08 = -20.88 + -0.19)

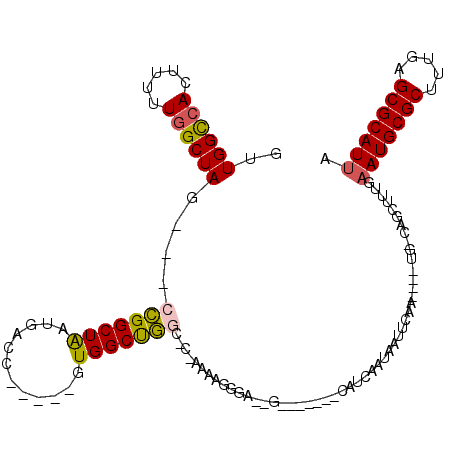
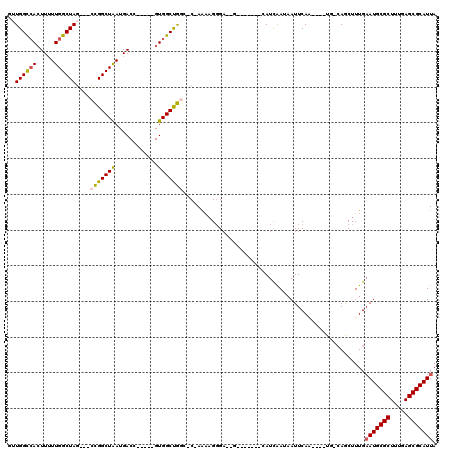
| Location | 2,098,005 – 2,098,101 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2098005 96 + 22407834 UUAUUGAUUCUCCCUUUU-GGCCAGCCACUGUCAUCAGCCGGCUAGCCAAAAAGUGGCCAACGAAACUCUGGAGGCGAAAUA-GCCCCCAA----AACUGAA ...(((.((..(((((((-(((.((((.(((....)))..)))).))).))))).))..))))).....(((.(((......-))).))).----....... ( -28.40) >DroSec_CAF1 115096 96 + 1 UUAUCGAUGCUCCCUUUU-GGCCAGCCACGGUCAUUAGCCGGCUAGCCAAAAAGUGGCCAACGAAACUCUGGAGGCGAAAGA-GCCCCCAA----AACUGAA ...(((..((..(.((((-(((.((((..(((.....))))))).))))))).)..))...))).....(((.(((......-))).))).----....... ( -32.80) >DroSim_CAF1 114964 96 + 1 UUAUUGAUGCUCCCUUUU-GGCCAGCCACGGUCAUUAGCCGGCUAGCCAAAAAGUGGCCAACGAAACUCUGGAGGCGAAAGA-GCCCCCAA----AACUGAA ...(((..((..(.((((-(((.((((..(((.....))))))).))))))).)..))...))).....(((.(((......-))).))).----....... ( -30.70) >DroEre_CAF1 117714 79 + 1 UUAUUGAUGCUCCCUUUU-GGUCAGCCACGGUCAUUAGCCGGCUAGCCAAAAAGUUGCCAAGAC-----------------A-GCCUUUGA----AACUGGA ..........(((.((((-((..((((..(((.....)))))))..))))))((((..((((..-----------------.-...)))).----))))))) ( -20.40) >DroYak_CAF1 119852 87 + 1 UUAUUGAUGCUCCCUUUUCGGCCAGCCACGGUCAUUAGCCGGCUAGCCAA-AAGUGGCCAACGAAGC--------------AGGCCUUCCAACUGAACUGAA ...(((.((..(((((((.(((.((((..(((.....))))))).)))))-))).)).)).)))..(--------------((..(........)..))).. ( -22.70) >consensus UUAUUGAUGCUCCCUUUU_GGCCAGCCACGGUCAUUAGCCGGCUAGCCAAAAAGUGGCCAACGAAACUCUGGAGGCGAAA_A_GCCCCCAA____AACUGAA ...(((.((.((.(((((.(((.((((..(((.....))))))).))).))))).)).)).)))...................................... (-16.34 = -17.06 + 0.72)

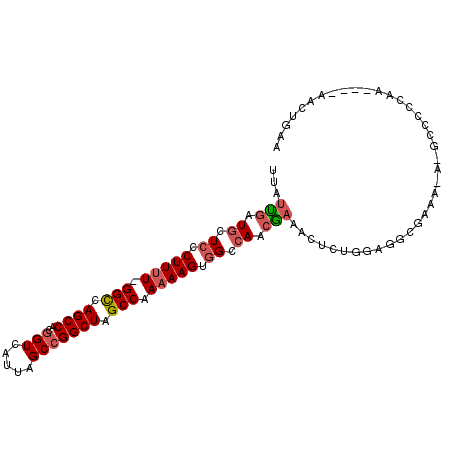
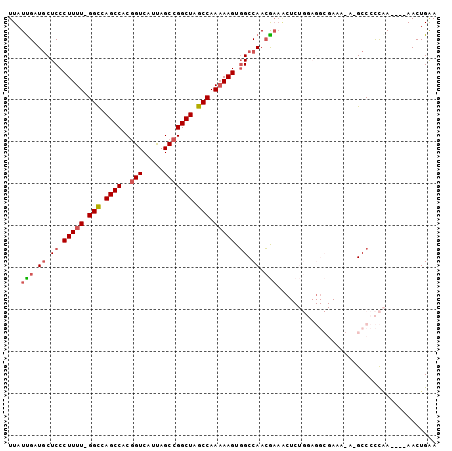
| Location | 2,098,005 – 2,098,101 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -21.78 |
| Energy contribution | -25.22 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2098005 96 - 22407834 UUCAGUU----UUGGGGGC-UAUUUCGCCUCCAGAGUUUCGUUGGCCACUUUUUGGCUAGCCGGCUGAUGACAGUGGCUGGCC-AAAAGGGAGAAUCAAUAA .....((----((((((((-......))))))))))....((((.(..(((((((((((((((.(((....))))))))))))-))))))..)...)))).. ( -44.40) >DroSec_CAF1 115096 96 - 1 UUCAGUU----UUGGGGGC-UCUUUCGCCUCCAGAGUUUCGUUGGCCACUUUUUGGCUAGCCGGCUAAUGACCGUGGCUGGCC-AAAAGGGAGCAUCGAUAA .....((----((((((((-......))))))))))..(((...((..(((((((((((((((((....).))..))))))))-))))))..))..)))... ( -45.80) >DroSim_CAF1 114964 96 - 1 UUCAGUU----UUGGGGGC-UCUUUCGCCUCCAGAGUUUCGUUGGCCACUUUUUGGCUAGCCGGCUAAUGACCGUGGCUGGCC-AAAAGGGAGCAUCAAUAA .....((----((((((((-......))))))))))....((((((..(((((((((((((((((....).))..))))))))-))))))..))..)))).. ( -43.30) >DroEre_CAF1 117714 79 - 1 UCCAGUU----UCAAAGGC-U-----------------GUCUUGGCAACUUUUUGGCUAGCCGGCUAAUGACCGUGGCUGACC-AAAAGGGAGCAUCAAUAA ..(((((----.....)))-)-----------------).....((..((((((((.((((((((....).))..))))).))-))))))..))........ ( -24.50) >DroYak_CAF1 119852 87 - 1 UUCAGUUCAGUUGGAAGGCCU--------------GCUUCGUUGGCCACUU-UUGGCUAGCCGGCUAAUGACCGUGGCUGGCCGAAAAGGGAGCAUCAAUAA ....((((........((((.--------------((...)).))))..((-(((((((((((((....).))..))))))))))))...))))........ ( -32.10) >consensus UUCAGUU____UUGGGGGC_U_UUUCGCCUCCAGAGUUUCGUUGGCCACUUUUUGGCUAGCCGGCUAAUGACCGUGGCUGGCC_AAAAGGGAGCAUCAAUAA ............(((((((.......))))))).......((((((..(((((((((((((((.(........)))))))))).))))))..))..)))).. (-21.78 = -25.22 + 3.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:59 2006