
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,285,190 – 20,285,280 |
| Length | 90 |
| Max. P | 0.704700 |

| Location | 20,285,190 – 20,285,280 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -18.22 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
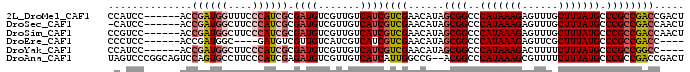
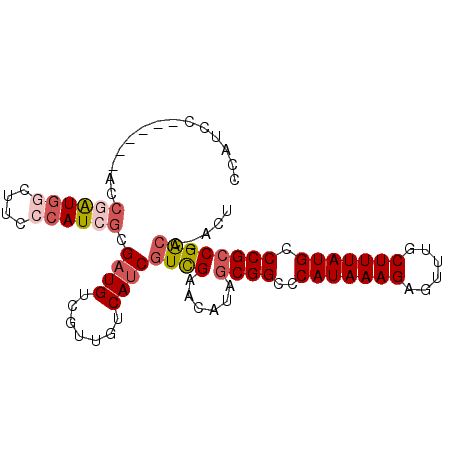
>2L_DroMel_CAF1 20285190 90 + 22407834 CCAUCC------ACCGAUGGUUUCCCAUCGCGAUGUCGUUGUCAUCGUCGAACAUAGCGGCCCAUAAAGAGUUUGCUUUAUGCCCGCCGACCGACU .((((.------..((((((....)))))).)))).....(((...((((......((((..(((((((......))))))).)))))))).))). ( -27.10) >DroSec_CAF1 35900 89 + 1 -CAUCC------ACCGAUGGCUUCCCAUCGCGAUGUCGUUGUCAUCGUCGAACAUAGCGGCCCAUAAAGAGUUUGCUUUAUGCCCGCCGACCAACU -((((.------..((((((....)))))).))))..((((.....((((......((((..(((((((......))))))).)))))))))))). ( -26.10) >DroSim_CAF1 34768 90 + 1 CCGUCC------ACCGAUGGCUUCCCAUCGCGAUGUCGUUGUCAUCGUCGAACAUAGCGGCCCAUAAAGAGUUUGCUUUAUGCCCGCCGACCAACU .((((.------..((((((....)))))).))))..((((.....((((......((((..(((((((......))))))).)))))))))))). ( -26.00) >DroEre_CAF1 21031 82 + 1 CCCUCC------ACCGAUGGC----GAUGUCGUUGUCAUCGUCAUCGUCGAACAUAGCGGCCCAUAAAGAGUUCGCUUUAUGCCCGCCGACC---- ......------...((((((----((((.......))))))))))((((......((((..(((((((......))))))).)))))))).---- ( -29.80) >DroYak_CAF1 16249 86 + 1 CCAUCC------ACCGAUGGCUUCCCAUCGCGAUGUCGUUGUCAUCGUCGAACAUAGCGGCCCAUAAAGACUUUUCUUUAUGCCCGCCGGCC---- (((((.------...)))))....((.((((((((.......))))).))).....((((..((((((((....)))))))).)))).))..---- ( -27.60) >DroAna_CAF1 3835 94 + 1 UAGUCCCGGCAGUCCAGUGCCUUCCCAUCGAGAUGUCGUUGUCAUCAUUGGCCG--ACGGCCCAUAAAGCGUUUUCUUUAUGCCCGCCGACCGACU .((((.((((.((((.(((......))).).)))((((((((((....))).))--))))).(((((((......)))))))...))))...)))) ( -25.40) >consensus CCAUCC______ACCGAUGGCUUCCCAUCGCGAUGUCGUUGUCAUCGUCGAACAUAGCGGCCCAUAAAGAGUUUGCUUUAUGCCCGCCGACC_ACU ..............((((((....)))))).((((.......))))((((......((((..(((((((......))))))).))))))))..... (-18.22 = -19.30 + 1.08)
| Location | 20,285,190 – 20,285,280 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -22.85 |
| Energy contribution | -24.13 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
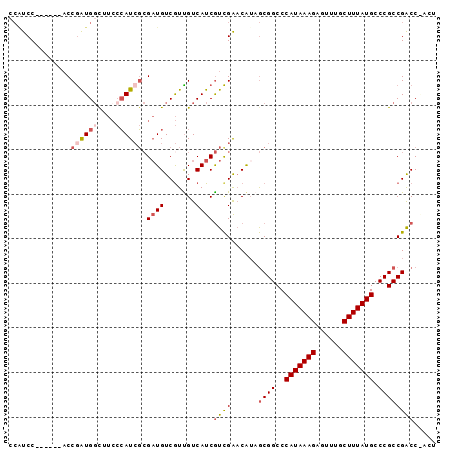
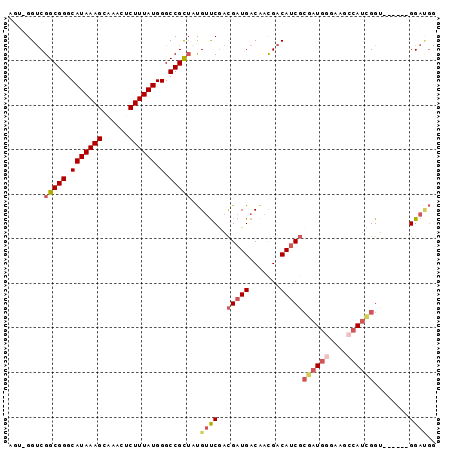
>2L_DroMel_CAF1 20285190 90 - 22407834 AGUCGGUCGGCGGGCAUAAAGCAAACUCUUUAUGGGCCGCUAUGUUCGACGAUGACAACGACAUCGCGAUGGGAAACCAUCGGU------GGAUGG .(((((..(((((.(((((((......)))))).).)))))....)))))...........((((.((((((....))))))..------.)))). ( -35.50) >DroSec_CAF1 35900 89 - 1 AGUUGGUCGGCGGGCAUAAAGCAAACUCUUUAUGGGCCGCUAUGUUCGACGAUGACAACGACAUCGCGAUGGGAAGCCAUCGGU------GGAUG- .((((((((.(((((((..(((...(((.....)))..)))))))))).))))..))))..((((.((((((....))))))..------.))))- ( -31.10) >DroSim_CAF1 34768 90 - 1 AGUUGGUCGGCGGGCAUAAAGCAAACUCUUUAUGGGCCGCUAUGUUCGACGAUGACAACGACAUCGCGAUGGGAAGCCAUCGGU------GGACGG ........(((((.(((((((......)))))).).))))).((((((.(((((.......)))))((((((....)))))).)------))))). ( -32.00) >DroEre_CAF1 21031 82 - 1 ----GGUCGGCGGGCAUAAAGCGAACUCUUUAUGGGCCGCUAUGUUCGACGAUGACGAUGACAACGACAUC----GCCAUCGGU------GGAGGG ----....(((((.(((((((......)))))).).))))).(.((((.(((((.(((((.......))))----).))))).)------))).). ( -28.40) >DroYak_CAF1 16249 86 - 1 ----GGCCGGCGGGCAUAAAGAAAAGUCUUUAUGGGCCGCUAUGUUCGACGAUGACAACGACAUCGCGAUGGGAAGCCAUCGGU------GGAUGG ----..(((((((.((((((((....))))))).).)))))..(((((.(((((.......)))))((((((....)))))).)------)))))) ( -32.20) >DroAna_CAF1 3835 94 - 1 AGUCGGUCGGCGGGCAUAAAGAAAACGCUUUAUGGGCCGU--CGGCCAAUGAUGACAACGACAUCUCGAUGGGAAGGCACUGGACUGCCGGGACUA (((((((((((((.(((((((......)))))).).))))--)))))........((.(((....))).))....((((......))))..)))). ( -35.30) >consensus AGU_GGUCGGCGGGCAUAAAGCAAACUCUUUAUGGGCCGCUAUGUUCGACGAUGACAACGACAUCGCGAUGGGAAGCCAUCGGU______GGAUGG ........(((((.(((((((......)))))).).)))))..((((..(((((.......)))))((((((....))))))........)))).. (-22.85 = -24.13 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:32 2006