| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,280,191 – 20,280,307 |
| Length | 116 |
| Max. P | 0.994910 |

| Location | 20,280,191 – 20,280,307 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20280191 116 + 22407834 CCAUUUUCACUUCCACUUCCGCCGCAC----ACUCACACACACAUGCACACUACUAGGACGUGCAUGCAUAUAUAUCCAUACACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ...........................----..(((.(((.((((..........(((((...(((((((((.(((........))).))))))))).))))).....))))))).))). ( -23.96) >DroSec_CAF1 30949 118 + 1 CCAUUUUCACUUCCACUUCCGCCGCACUCACACACACACACACAUGCACACUACUAGGACGUGCAUGCAUA--UAUCAAUACACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ..................................((.(((.((((..........(((((...((((((((--(..............))))))))).))))).....))))))).)).. ( -21.00) >DroSim_CAF1 29797 116 + 1 CCAUUUUCACUUCCACUUCCGCCGCACUCACACAC--ACACACAUGCACACUACUAGGACGUGCAUGCAUA--UAUCAAUACACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ................................((.--(((.((((..........(((((...((((((((--(..............))))))))).))))).....))))))).)).. ( -21.00) >DroEre_CAF1 15981 102 + 1 CCAUUUUCACUUCCACUUCCGCCGCAC----------ACACACAGGCGCACUAC-AGGAUGUGCCUGCAUA--UAC-----CACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ...........................----------.....((((((((....-(((((...(.((((((--(..-----.......))))))).).)))))......)).)))))).. ( -19.00) >DroYak_CAF1 11218 101 + 1 CCAUUUUCACUUCCACUUCCGCCGCAC----------ACACACAUACACACUACUAGGAUGUGCCUGCAU----AC-----UACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ...........................----------.((.(((.(((.......(((((...(.(((((----(.-----........)))))).).)))))......)))))).)).. ( -14.32) >consensus CCAUUUUCACUUCCACUUCCGCCGCAC____AC_C__ACACACAUGCACACUACUAGGACGUGCAUGCAUA__UAUC_AUACACAUACAUAUGCAUGAGUCCUUCACAAUGUUGUCUGAA ......................................((.(((.(((.......(((((...((((((((..................)))))))).)))))......)))))).)).. (-17.03 = -17.07 + 0.04)


| Location | 20,280,191 – 20,280,307 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
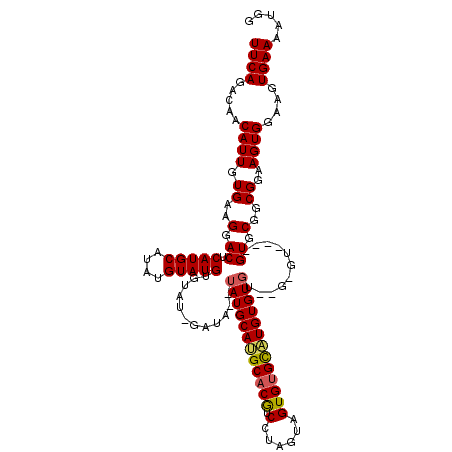
>2L_DroMel_CAF1 20280191 116 - 22407834 UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUGUAUGGAUAUAUAUGCAUGCACGUCCUAGUAGUGUGCAUGUGUGUGUGAGU----GUGCGGCGGAAGUGGAAGUGAAAAUGG .........((((.....(.(((((((((((((((((((((....)))))))))(((((((.(......))))))))))))))))))))----.).(.((....)).).......)))). ( -35.40) >DroSec_CAF1 30949 118 - 1 UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUGUAUUGAUA--UAUGCAUGCACGUCCUAGUAGUGUGCAUGUGUGUGUGUGUGUGAGUGCGGCGGAAGUGGAAGUGAAAAUGG ((((.....((((.((..(.(((((((((((..((((((((....))--)))(((((((((.(......)))))))))))))..))))))))))).)..))..))))....))))..... ( -36.90) >DroSim_CAF1 29797 116 - 1 UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUGUAUUGAUA--UAUGCAUGCACGUCCUAGUAGUGUGCAUGUGUGU--GUGUGUGAGUGCGGCGGAAGUGGAAGUGAAAAUGG ((((.....((((.((..(.(((((((((((..(((((((....)))--)))(((((((((.(......)))))))))))..)--)))))))))).)..))..))))....))))..... ( -35.60) >DroEre_CAF1 15981 102 - 1 UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUG-----GUA--UAUGCAGGCACAUCCU-GUAGUGCGCCUGUGUGU----------GUGCGGCGGAAGUGGAAGUGAAAAUGG ((((.(......).))))....((((...(((...(((.-----(((--(((((((((.(((...-...))).))))))..))----------)))).)))...)))...))))...... ( -27.70) >DroYak_CAF1 11218 101 - 1 UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUA-----GU----AUGCAGGCACAUCCUAGUAGUGUGUAUGUGUGU----------GUGCGGCGGAAGUGGAAGUGAAAAUGG .........((((..........((((((((((((..((-----.(----((..(((.....))).))).))..)))))))))----------)))(.((....)).).......)))). ( -25.10) >consensus UUCAGACAACAUUGUGAAGGACUCAUGCAUAUGUAUGUGUAU_GAUA__UAUGCAUGCACGUCCUAGUAGUGUGCAUGUGUGU__G_GU____GUGCGGCGGAAGUGGAAGUGAAAAUGG ((((.....((((.((..(.((.(((((....)))))............((((((((((((.(......)))))))))))))...........)).)..))..))))....))))..... (-22.48 = -22.44 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:30 2006