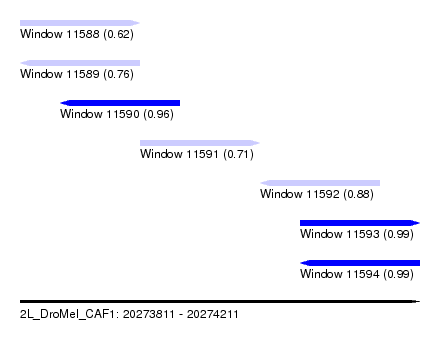
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,273,811 – 20,274,211 |
| Length | 400 |
| Max. P | 0.988709 |

| Location | 20,273,811 – 20,273,931 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.24 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273811 120 + 22407834 UUGCUGUGCAGCUGAAAAUCUGGAAAAAAGGAGAGGAGUGUUUAAAUAUUCGCCCUGCACUUCAGUCGUGCCUUAAAGGAAAGCCCUUUAAAGCGAAGAAGAUGAAAAUGCAGGCCACCC ..(((....)))........(((............((((((....))))))..((((((.((((.((.(((.(((((((.....))))))).)))..))...))))..)))))))))... ( -30.00) >DroSec_CAF1 24665 120 + 1 UUGCUGUGCAGCUGAAAAUCUGGGAAAAAAGAGAGGAGUGUUUAAAUAUUCGCCCUGCACUUCAGUCGUGCCUUAAAGGAAAAACCUUUAAAGCGAAGAAGAUGAAAAUGCAGGCCACCC ..(((....))).........(((...........((((((....))))))(.((((((.((((.((.(((.(((((((.....))))))).)))..))...))))..)))))).).))) ( -31.90) >DroSim_CAF1 23542 119 + 1 UUGCUGUGCAGCUGAAAAUCUG-GAAAAAAGAGAGGAGUGUUUAAAUAUUCGCCCUGCACUUCAGUCGUGCCUUAAAGGAAAACCCUUUAAAGCGAGGAAGAUGAAAAUGCAGGCCACCC ..(((....)))........((-(...........((((((....))))))..((((((.((((.((.(((.(((((((.....))))))).)))..))...))))..)))))))))... ( -30.90) >DroEre_CAF1 4804 110 + 1 UUGCUGUGCAGCUGAAAAUCUGG-AA-AAAGAGAGGAGCGUUUAAAUAUUCGCCCUGCACUUCAGCCGUGCCUUAAAGGAAAACCUUUAAAA----AGACG----AAGUGCAGGCCACCC ...(((..(.((((((..(((..-..-..))).(((.(((..........))))))....))))))(((...(((((((....)))))))..----..)))----..)..)))....... ( -29.80) >DroYak_CAF1 5252 111 + 1 UUGCUGUGCAGCUGAAAAUCUGG-AAAAAAGAGAGGAGUGUUUAAAUAUUCGCCCUGCACUUCAGCUGUGCCUUAAAGGAAAACCCUUUAAA----AGAAG----AAAUGCAGGCCACCC ((((.(..((((((((..(((..-.....))).(((.(((..........))))))....))))))))..).(((((((.....))))))).----.....----....))))....... ( -30.30) >consensus UUGCUGUGCAGCUGAAAAUCUGG_AAAAAAGAGAGGAGUGUUUAAAUAUUCGCCCUGCACUUCAGUCGUGCCUUAAAGGAAAACCCUUUAAAGCGAAGAAGAUGAAAAUGCAGGCCACCC ((((.(..(.((((((..(((........))).(((.(((..........))))))....)))))).)..).(((((((.....)))))))..................))))....... (-24.76 = -24.24 + -0.52)



| Location | 20,273,811 – 20,273,931 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.29 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273811 120 - 22407834 GGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGGCUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAACACUCCUCUCCUUUUUUCCAGAUUUUCAGCUGCACAGCAA ...((.((((((((((..((......((((((((((.....))))))))))..))..)))))))))).))......................................(((....))).. ( -31.20) >DroSec_CAF1 24665 120 - 1 GGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGUUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAACACUCCUCUCUUUUUUCCCAGAUUUUCAGCUGCACAGCAA (((.(.((((((((((..((......((((((((((.....))))))))))..))..)))))))))).)((...............))........))).........(((....))).. ( -33.16) >DroSim_CAF1 23542 119 - 1 GGGUGGCCUGCAUUUUCAUCUUCCUCGCUUUAAAGGGUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAACACUCCUCUCUUUUUUC-CAGAUUUUCAGCUGCACAGCAA ...((.((((((((((..((......((((((((((.....))))))))))..))..)))))))))).))...........................-..........(((....))).. ( -31.20) >DroEre_CAF1 4804 110 - 1 GGGUGGCCUGCACUU----CGUCU----UUUUAAAGGUUUUCCUUUAAGGCACGGCUGAAGUGCAGGGCGAAUAUUUAAACGCUCCUCUCUUU-UU-CCAGAUUUUCAGCUGCACAGCAA (((.(((((((((((----((((.----(((((((((....)))))))))...))).))))))))))(((..........))).)).)))...-..-...........(((....))).. ( -36.60) >DroYak_CAF1 5252 111 - 1 GGGUGGCCUGCAUUU----CUUCU----UUUAAAGGGUUUUCCUUUAAGGCACAGCUGAAGUGCAGGGCGAAUAUUUAAACACUCCUCUCUUUUUU-CCAGAUUUUCAGCUGCACAGCAA ...((.(((((((((----(..((----((((((((.....))))))))....))..)))))))))).))..........................-...........(((....))).. ( -28.10) >consensus GGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGGUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAACACUCCUCUCUUUUUU_CCAGAUUUUCAGCUGCACAGCAA (((((.((((((((((..........((((((((((.....))))))))))......)))))))))).............))))).......................(((....))).. (-24.01 = -25.29 + 1.28)

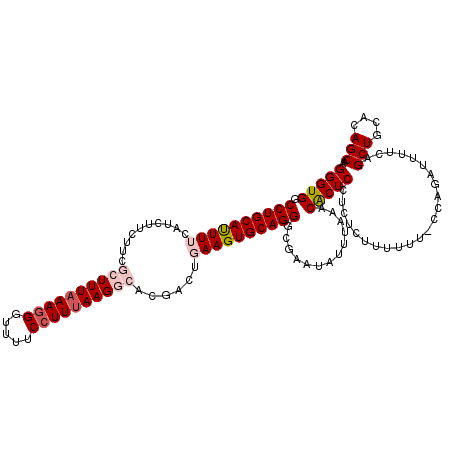
| Location | 20,273,851 – 20,273,971 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -28.57 |
| Energy contribution | -30.21 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273851 120 - 22407834 CCUUUCCAACUGUUUUGCUUUUACCUCGGUUUCCAGCGGAGGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGGCUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAA .............(((((.....((((.((.....)).))))....((((((((((..((......((((((((((.....))))))))))..))..)))))))))))))))........ ( -38.10) >DroSec_CAF1 24705 120 - 1 CCUUUCCAACUGUUUUGCUUUUACCUCGGUUUCCAGCGGAGGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGUUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAA .............(((((.....((((.((.....)).))))....((((((((((..((......((((((((((.....))))))))))..))..)))))))))))))))........ ( -38.40) >DroSim_CAF1 23581 120 - 1 CCUUUCCAACUGUUUUGCUUUUACCUCGGUUUCCAGCGGAGGGUGGCCUGCAUUUUCAUCUUCCUCGCUUUAAAGGGUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAA .............(((((((((((.((((((....(.(((((((((.........))))))))).)((((((((((.....))))))))))..)))))).))).))))))))........ ( -38.50) >DroEre_CAF1 4842 112 - 1 CCUUUCCAACUGUUUUGCUUUUAGCUCGGUUUUCCGGGGAGGGUGGCCUGCACUU----CGUCU----UUUUAAAGGUUUUCCUUUAAGGCACGGCUGAAGUGCAGGGCGAAUAUUUAAA (((..(((((((....((.....)).)))))....))..))).((.(((((((((----((((.----(((((((((....)))))))))...))).)))))))))).)).......... ( -43.80) >DroYak_CAF1 5291 112 - 1 CCUUUCCAACUGUUUUGCUUUUACCUCGGUUUUCAGGGGAGGGUGGCCUGCAUUU----CUUCU----UUUAAAGGGUUUUCCUUUAAGGCACAGCUGAAGUGCAGGGCGAAUAUUUAAA (((((((..(((....(((........)))...))))))))))((.(((((((((----(..((----((((((((.....))))))))....))..)))))))))).)).......... ( -36.30) >consensus CCUUUCCAACUGUUUUGCUUUUACCUCGGUUUCCAGCGGAGGGUGGCCUGCAUUUUCAUCUUCUUCGCUUUAAAGGGUUUUCCUUUAAGGCACGACUGAAGUGCAGGGCGAAUAUUUAAA .((((((..(((....(((........)))...))).))))))((.((((((((((..........((((((((((.....))))))))))......)))))))))).)).......... (-28.57 = -30.21 + 1.64)


| Location | 20,273,931 – 20,274,051 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273931 120 + 22407834 UCCGCUGGAAACCGAGGUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGGAAAUAGAU (((((((....)...((((((((.....((((((..........))))))....)))))))))))..((((((((((...(((.(......).))).)))).))))))..)))....... ( -30.50) >DroSec_CAF1 24785 120 + 1 UCCGCUGGAAACCGAGGUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGGAAAUAGAU (((((((....)...((((((((.....((((((..........))))))....)))))))))))..((((((((((...(((.(......).))).)))).))))))..)))....... ( -30.50) >DroSim_CAF1 23661 120 + 1 UCCGCUGGAAACCGAGGUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGCAAAUAGAU (((((((....)...((((((((.....((((((..........))))))....)))))))))))....((((((((...(((.(......).))).)))).)))))))........... ( -29.30) >DroEre_CAF1 4914 120 + 1 UCCCCGGAAAACCGAGCUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGGAAAUAGAU ....(((....)))..(((...((((((((((((..........)))))).....))))))...(((((((((((((...(((.(......).))).)))).)))).)))))...))).. ( -25.90) >DroYak_CAF1 5363 120 + 1 UCCCCUGAAAACCGAGGUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGGAAAUAGAU ....(((........((((((((.....((((((..........))))))....))))))))..(((((((((((((...(((.(......).))).)))).)))).)))))...))).. ( -29.00) >consensus UCCGCUGGAAACCGAGGUAAAAGCAAAACAGUUGGAAAGGAAAGCAGCUGAAAACUUUUGCCAGCCAUUUGCCGGUGAAAAUUGAGAUUUAUGAGUGCACUAGGUAGGAUGGAAAUAGAU ...............((((((((.....((((((..........))))))....))))))))..(((((((((((((...(((.(......).))).)))).)))).)))))........ (-25.92 = -26.32 + 0.40)

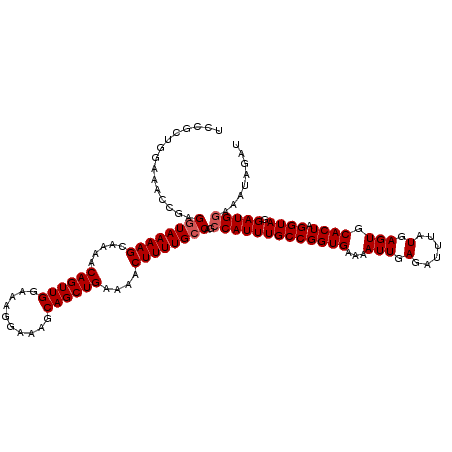
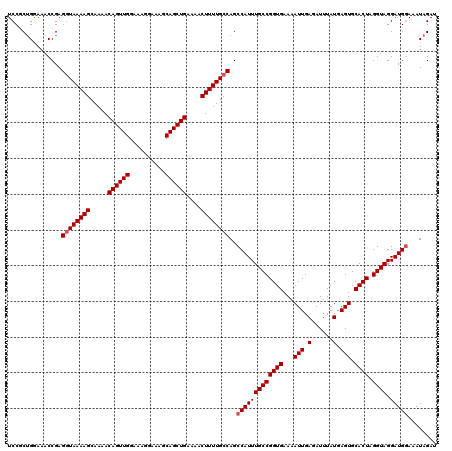
| Location | 20,274,051 – 20,274,171 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20274051 120 - 22407834 CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACCCUUUCUGUUUCUCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... ( -31.30) >DroSec_CAF1 24905 120 - 1 CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACUCUUUCUGUUUCCCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... ( -31.30) >DroSim_CAF1 23781 120 - 1 CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACCCUUUCUGUUUCCCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... ( -31.30) >DroEre_CAF1 5034 120 - 1 CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACCCUUUCUGUUUCUCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... ( -31.30) >DroYak_CAF1 5483 120 - 1 CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACCCUUUCUGUUUCUCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... ( -31.30) >consensus CAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAGCCGGGCUUUUGUGUGCGCUCCUUACCCUUUCUGUUUCUCU ........(((((......((((.((((.....((((((((....))))))))...(((((((((((............))))))..))))).....))))))))......))))).... (-31.30 = -31.30 + 0.00)



| Location | 20,274,091 – 20,274,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20274091 120 + 22407834 CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). ( -31.40) >DroSec_CAF1 24945 120 + 1 CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). ( -31.40) >DroSim_CAF1 23821 120 + 1 CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). ( -31.40) >DroEre_CAF1 5074 120 + 1 CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). ( -31.40) >DroYak_CAF1 5523 120 + 1 CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). ( -31.40) >consensus CUGCAUCUAUUUUGCCGAGUUUGUUUUUGCCAAGUUGGAACUUGGCAUAUUUCUCCAUUACGAAUGGCGCUUGCAAAUUGAUUUGAUUUGAUUCGAUUUUGAAGUCGAUAAGCCCGCUCU ..(((.......))).(((...((...((((((((....)))))))).))..)))......((.(((.(((((((((((.....))))))..((((((....)))))))))))))).)). (-31.40 = -31.40 + 0.00)

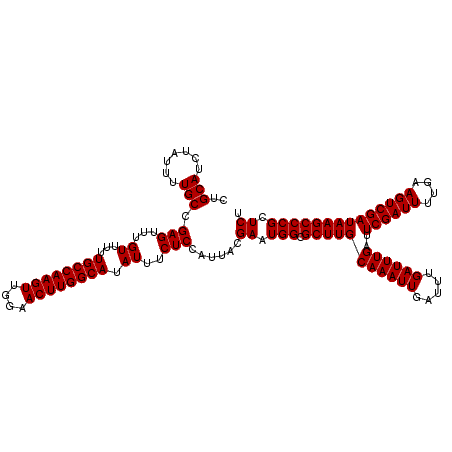
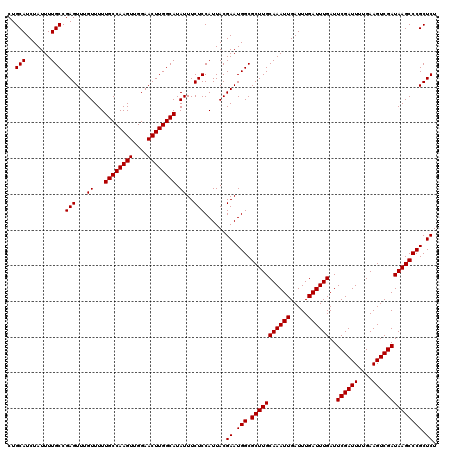
| Location | 20,274,091 – 20,274,211 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20274091 120 - 22407834 AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. ( -26.80) >DroSec_CAF1 24945 120 - 1 AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. ( -26.80) >DroSim_CAF1 23821 120 - 1 AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. ( -26.80) >DroEre_CAF1 5074 120 - 1 AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. ( -26.80) >DroYak_CAF1 5523 120 - 1 AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. ( -26.80) >consensus AGAGCGGGCUUAUCGACUUCAAAAUCGAAUCAAAUCAAAUCAAUUUGCAAGCGCCAUUCGUAAUGGAGAAAUAUGCCAAGUUCCAACUUGGCAAAAACAAACUCGGCAAAAUAGAUGCAG ...(((.((((.((((........))))..(((((.......))))).))))(((.(((........)))...((((((((....))))))))...........)))........))).. (-26.80 = -26.80 + 0.00)

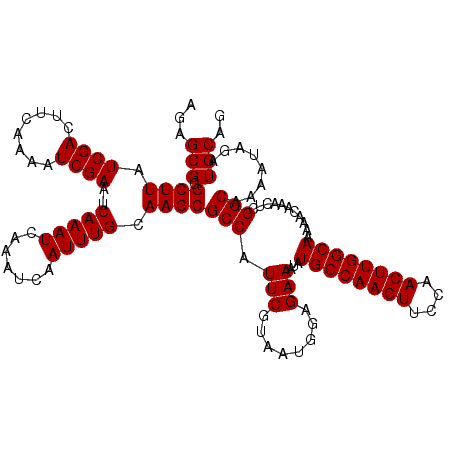
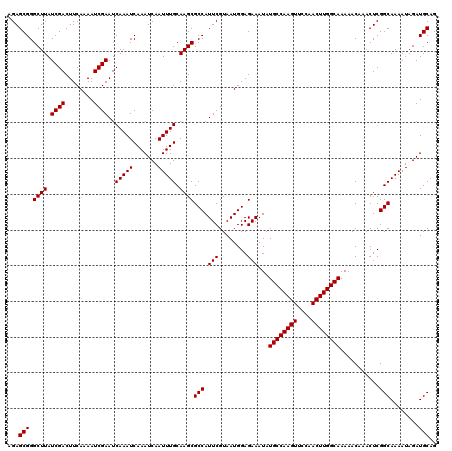
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:21 2006