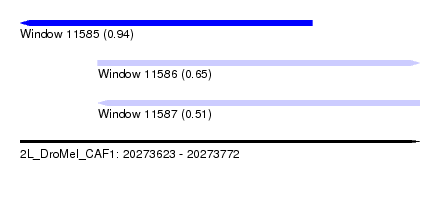
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,273,623 – 20,273,772 |
| Length | 149 |
| Max. P | 0.939315 |

| Location | 20,273,623 – 20,273,732 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273623 109 - 22407834 ACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACACCUCCUCCGAAACCCAUCGCCGA----------- ...(.((.(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))))...............((......)))).).----------- ( -20.20) >DroSec_CAF1 24476 109 - 1 ACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACUCCUCCUCCGAAACCCAUCGCCGA----------- ...(.((.(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))))...............((......)))).).----------- ( -20.20) >DroSim_CAF1 23354 109 - 1 ACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACUCCUCCUCCGAAACCCAUCGCCGA----------- ...(.((.(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))))...............((......)))).).----------- ( -20.20) >DroEre_CAF1 4608 117 - 1 ACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACU---CCUCCGAAACCCAUCCCCGAGCCCCGAGUCC ........(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))))....(((---((((.(..........).)))....)))).. ( -21.00) >DroYak_CAF1 5067 106 - 1 ACACCGCACAGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACU---CCUCCGAAACCCAUCUUCGA----------- ..........(((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))).........---....((((.......)))).----------- ( -17.40) >consensus ACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGCCCCACUCCUCCUCCGAAACCCAUCGCCGA___________ ........(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))))......................................... (-17.86 = -18.06 + 0.20)



| Location | 20,273,652 – 20,273,772 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273652 120 + 22407834 GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCAGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((((....((..((((((((((((...((....)).)))))).....))))))..))....))))))))(((.....)))..((((..((....))..)))).............. ( -27.20) >DroSec_CAF1 24505 120 + 1 GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCAGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((((....((..((((((((((((...((....)).)))))).....))))))..))....))))))))(((.....)))..((((..((....))..)))).............. ( -27.20) >DroSim_CAF1 23383 120 + 1 GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCAGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((((....((..((((((((((((...((....)).)))))).....))))))..))....))))))))(((.....)))..((((..((....))..)))).............. ( -27.20) >DroEre_CAF1 4645 120 + 1 GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCAGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((((....((..((((((((((((...((....)).)))))).....))))))..))....))))))))(((.....)))..((((..((....))..)))).............. ( -27.20) >DroYak_CAF1 5093 120 + 1 GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCUGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((.((.((.((.((((((((((((...((....)).)))))).....))))))....))...)).)).))))))........((((..((....))..)))).............. ( -26.00) >consensus GCUGCAAAGGGAUUCGGUUUUAUGGCGGAAAUGGCAACGCUUGCCACAAAAUAAAACAAAAGAUAUUUGCAGUGCGGUGUGUGCGACGAUAAAGUAGACUAGGUCGAAAUAAAGAUAAAA ((((((((....((..((((((((((((...((....)).)))))).....))))))..))....))))))))(((.....)))..((((..((....))..)))).............. (-26.16 = -26.36 + 0.20)

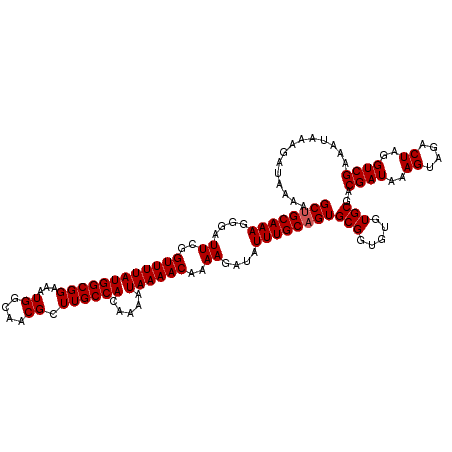
| Location | 20,273,652 – 20,273,772 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273652 120 - 22407834 UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......)).(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))))). ( -21.40) >DroSec_CAF1 24505 120 - 1 UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......)).(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))))). ( -21.40) >DroSim_CAF1 23383 120 - 1 UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......)).(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))))). ( -21.40) >DroEre_CAF1 4645 120 - 1 UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......)).(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))))). ( -21.40) >DroYak_CAF1 5093 120 - 1 UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACAGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......))...(((((....(((.(((((((...((((((.....)))))).......))))))).)))....)))))... ( -18.20) >consensus UUUUAUCUUUAUUUCGACCUAGUCUACUUUAUCGUCGCACACACCGCACUGCAAAUAUCUUUUGUUUUAUUUUGUGGCAAGCGUUGCCAUUUCCGCCAUAAAACCGAAUCCCUUUGCAGC ...............(((.(((......)))..)))((.......)).(((((((....(((.(((((((...((((((.....)))))).......))))))).)))....))))))). (-20.36 = -20.56 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:15 2006