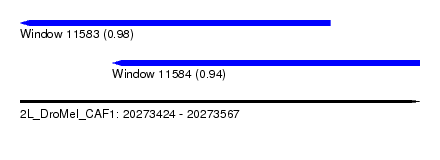
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,273,424 – 20,273,567 |
| Length | 143 |
| Max. P | 0.978270 |

| Location | 20,273,424 – 20,273,535 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273424 111 - 22407834 GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC------CGUCCGGCAACAGCAUUUGCACAUGACCAUAGA--- ..((((((.....((((((............))))))......................((((((.....))))))..------..))))))....((....)).............--- ( -23.40) >DroSec_CAF1 24275 114 - 1 GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC------CGUCCGGCAACAGCAUUUGCACAUGACCGUAGAACA ..((((((.....((((((............))))))......................((((((.....))))))..------..)))))).......(((((........)))))... ( -23.80) >DroSim_CAF1 23153 114 - 1 GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC------CGUCCGGCAGCAGCAUUUGCACAUGACCGUAGAACA ..((((((.....((((((............))))))......................((((((.....))))))..------..)))))).......(((((........)))))... ( -23.80) >DroEre_CAF1 4399 117 - 1 GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCCUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUUUCGUUCCCGUCCGGCAACAGCAUUUGCACAUGACCGUAGA--- ..((((((.....(((.(((.....((......))......................((((((((.....)))))))).)))))).))))))....((....)).............--- ( -27.10) >DroYak_CAF1 4876 111 - 1 GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCU------CGUCCGGCAACAGCAUUUGCACAUGACCGUAGA--- ..((((((.....((((((............))))))......................((((((.....))))))..------..))))))....((....)).............--- ( -23.40) >consensus GAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC______CGUCCGGCAACAGCAUUUGCACAUGACCGUAGA___ ..((((((.....((((((............))))))......................((((((.....))))))..........))))))....((....))................ (-22.98 = -23.18 + 0.20)

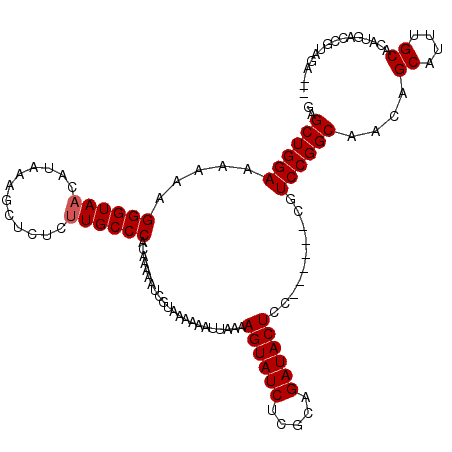
| Location | 20,273,457 – 20,273,567 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.43 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20273457 110 - 22407834 UAAACGGCACAGAAACAAAAAAAAAAAAUGGUGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC-- ...((((..(((..((..............))...))).......((((((............)))))).......))))...........((((((.....))))))..-- ( -15.74) >DroSec_CAF1 24311 109 - 1 UAAACGGCACAGAAACAAAAAA-AAACAUGGUGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC-- ...((((..(((..((......-.......))...))).......((((((............)))))).......))))...........((((((.....))))))..-- ( -15.82) >DroSim_CAF1 23189 109 - 1 UAAACGGCACAGAAACAAAAAA-AAACAUGGUGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC-- ...((((..(((..((......-.......))...))).......((((((............)))))).......))))...........((((((.....))))))..-- ( -15.82) >DroEre_CAF1 4436 106 - 1 UAAACGGCACAGAAAAA------AAUAAUGGUGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCCUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUUUCG .....((((........------......((.(((((...................))))).)))))).....................((((((((.....)))))))).. ( -18.84) >DroYak_CAF1 4909 102 - 1 UAAACGGCACAGAAAAA------A--AAUGGGGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCU-- .....((((........------.--...((((((((...................)))))))))))).......................((((((.....))))))..-- ( -18.81) >consensus UAAACGGCACAGAAACAAAAAA_AAAAAUGGUGAGCUGGAAAAAAGGGUAACAUAAAGCUCUCUUGCCCACAAAAAUCGUAAAAAAUUAAAAGUAUCUCGCAGAUACUCC__ .....((((....................((.(((((...................))))).)))))).......................((((((.....)))))).... (-15.32 = -15.16 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:12 2006