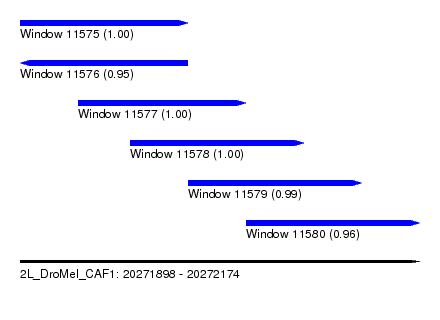
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,271,898 – 20,272,174 |
| Length | 276 |
| Max. P | 0.999923 |

| Location | 20,271,898 – 20,272,014 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.51 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271898 116 + 22407834 UUUAUAUGCAUGACAUGGCAAAUCAUCGGACUGAUUUGUUUACCAAGUCCA----UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAU (((((.(((..(((.((((((((((......)))))))....))).)))((----.....))....))).))))).......(((((((((.......)))))))))............. ( -27.00) >DroSec_CAF1 22771 116 + 1 UUUAUAUGCAUGACGGGGCAAAUCAUCGGACUGAUUUGUUUACCAAGUCCA----UACUCUGCAGAGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAU ....((((...(((.((((((((((......)))))))))).....)))))----))....(((((.......((....)).(((((((((.......)))))))))..)))))...... ( -31.20) >DroSim_CAF1 21652 116 + 1 UUUAUAUGCAUGACGGGGCAAAUCAUCGGACUGAUUUGUUUACCAAGUCCA----UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAU ....((((...(((.((((((((((......)))))))))).....)))))----))....((((........((....)).(((((((((.......)))))))))...))))...... ( -28.70) >DroEre_CAF1 2954 120 + 1 UUUACAUGCAUGACAUGGCAAAUCAUCGGACUGAUUUGUUUACCCAGCCCAAGUCUACUAUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAU ......((((((((.((((((((((......)))))))..........))).)))....)))))..((.....((....)).(((((((((.......))))))))).....))...... ( -28.20) >DroYak_CAF1 3369 116 + 1 UUUAUAUGCAUGACAUAGCAAAUUAUCGGACUGAUUUGUUUACCAAGUCCA----UACUAUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAU (((((.(((.((.(((((.....(((.(((((.............))))))----)))))))))..))).))))).......(((((((((.......)))))))))............. ( -29.32) >consensus UUUAUAUGCAUGACAUGGCAAAUCAUCGGACUGAUUUGUUUACCAAGUCCA____UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAU (((((.(((.((.(((((.........(((((.............))))).......)))))))..))).))))).......(((((((((.......)))))))))............. (-27.63 = -27.51 + -0.12)

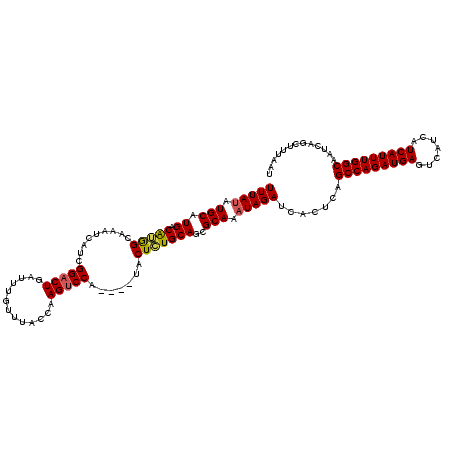
| Location | 20,271,898 – 20,272,014 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271898 116 - 22407834 AUUAAAGCUGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCGCUGCAGAGUA----UGGACUUGGUAAACAAAUCAGUCCGAUGAUUUGCCAUGUCAUGCAUAUAAA .......(((...((((((((..(((.(((((......))))).))))))))).))..))).(((----((.((.(((((((...(((.....)))..))))))).)))))))....... ( -34.00) >DroSec_CAF1 22771 116 - 1 AUUAAAGCAGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCUCUGCAGAGUA----UGGACUUGGUAAACAAAUCAGUCCGAUGAUUUGCCCCGUCAUGCAUAUAAA ......(((((..((.(((((..(((.(((((......))))).)))))))))))))))...(((----((((((.(((......))))))))((((........))))))))....... ( -32.00) >DroSim_CAF1 21652 116 - 1 AUUAAAGCAGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCGCUGCAGAGUA----UGGACUUGGUAAACAAAUCAGUCCGAUGAUUUGCCCCGUCAUGCAUAUAAA ......((((((.((((((((..(((.(((((......))))).))))))))).)).(((((.((----((((((.(((......)))))))).))).)))))...))).)))....... ( -32.40) >DroEre_CAF1 2954 120 - 1 AUUAAAGCUGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCGCUGCAUAGUAGACUUGGGCUGGGUAAACAAAUCAGUCCGAUGAUUUGCCAUGUCAUGCAUGUAAA ......(((((..((((((((..(((.(((((......))))).))))))))).))..(((.(((((((((((((((.........))))))))).).))))).)))))).))....... ( -33.40) >DroYak_CAF1 3369 116 - 1 AUUAAAGCUGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCGCUGCAUAGUA----UGGACUUGGUAAACAAAUCAGUCCGAUAAUUUGCUAUGUCAUGCAUAUAAA ......(((((..((((((((..(((.(((((......))))).))))))))).))..(((((((----((((((.(((......)))))))))......)))))))))).))....... ( -31.40) >consensus AUUAAAGCUGAUUGCCAAAUGAUGAUGACUCAUCUGGCUGAGUGAUCUAUUUGCGCUGCAGAGUA____UGGACUUGGUAAACAAAUCAGUCCGAUGAUUUGCCAUGUCAUGCAUAUAAA ......((((((.((((((((..(((.(((((......))))).))))))))).)).(((((.((....((((((.(((......))))))))).)).)))))...)))).))....... (-26.54 = -27.22 + 0.68)

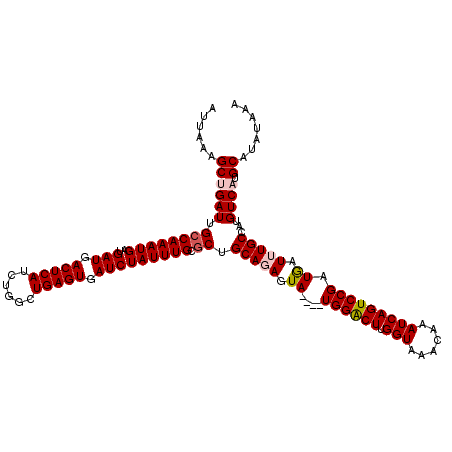

| Location | 20,271,938 – 20,272,054 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271938 116 + 22407834 UACCAAGUCCA----UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGGAACCAC .......(((.----.((((((.(((.......((....)).(((((((((.......))))))))).....))).......(((((((.......)))))))))))))...)))..... ( -34.70) >DroSec_CAF1 22811 116 + 1 UACCAAGUCCA----UACUCUGCAGAGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCAC .......((..----.((((((.((((((....((....)).(((((((((.......)))))))))....)))))).....(((((((.......)))))))))))))...))...... ( -34.80) >DroSim_CAF1 21692 116 + 1 UACCAAGUCCA----UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCAC .......((..----.((((((....(((....((....)).(((((((((.......)))))))))....)))........(((((((.......)))))))))))))...))...... ( -31.80) >DroEre_CAF1 2994 120 + 1 UACCCAGCCCAAGUCUACUAUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCGAACCAGAGUCCAGAAACCAC ............(((((...(((...)))..))))).((((.(((((((((.......)))))))))...............(((((((.......)))))))..))))........... ( -29.60) >DroYak_CAF1 3409 116 + 1 UACCAAGUCCA----UACUAUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCGAAAUCCGAGUCCAGAAACCAC ......(((((----.((((...(((.......((....)).(((((((((.......))))))))).....))).....)))).))).)).((...((.....))..)).......... ( -23.50) >consensus UACCAAGUCCA____UACUCUGCAGCGCAAAUAGAUCACUCAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCAC ................((((((....((.....((....)).(((((((((.......))))))))).....))........(((((((.......)))))))))))))........... (-26.44 = -26.92 + 0.48)



| Location | 20,271,974 – 20,272,094 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.42 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271974 120 + 22407834 CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGGAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......))))))))).....((((.((((.(((((((.......))))))).((((..((((.....))))..)))).)))).))))((..(((......)))..)). ( -35.50) >DroSec_CAF1 22847 120 + 1 CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......))))))))).....((((.((((.(((((((.......))))))).((((..(((.......)))..)))).)))).))))((..(((......)))..)). ( -32.30) >DroSim_CAF1 21728 120 + 1 CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCUGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......))))))))).....((((.((((.(((((((.......))))))).((((..(((.......)))..)))).)))).))))((..(((......)))..)). ( -32.30) >DroEre_CAF1 3034 120 + 1 CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCGAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......))))))))).....((((.((((.(((((((.......))))))).((((..(((.......)))..)))).)))).))))((..(((......)))..)). ( -32.20) >DroYak_CAF1 3445 120 + 1 CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCGAAAUCCGAGUCCAGAAACCACUUGCCCUCUCAAUUCGGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......)))))))))..((((..........(((((.((((...........)).)))))))......((((((.........))))))..))))............. ( -27.90) >consensus CAGCCAGAUGAGUCAUCAUCAUUUGGCAAUCAGCUUUAAUUAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCC ..(((((((((.......))))))))).....((((.((((.(((((((.......)))))))..(((..(((.......)))..)))..)))).))))((..(((......)))..)). (-30.70 = -30.42 + -0.28)


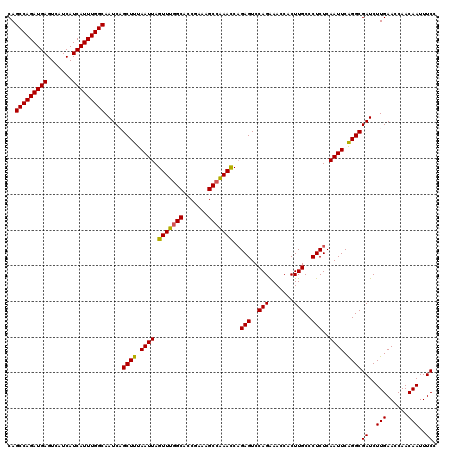
| Location | 20,272,014 – 20,272,134 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20272014 120 + 22407834 UAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGGAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....(((((((.(...((((((..((((..((((.....))))..)))).......(((((..(((......)))..).))))((((((...))))))......)))))).).))))))) ( -33.30) >DroSec_CAF1 22887 120 + 1 UAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....(((((((.(...((((((..((((..(((.......)))..)))).......(((((..(((......)))..).))))((((((...))))))......)))))).).))))))) ( -30.30) >DroSim_CAF1 21768 120 + 1 UAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....(((((((.(...((((((..((((..(((.......)))..)))).......(((((..(((......)))..).))))((((((...))))))......)))))).).))))))) ( -30.30) >DroEre_CAF1 3074 120 + 1 UAGUUUGGCACCGAAAGCCGAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....(((((((.(...((((((..((((..(((.......)))..)))).......(((((..(((......)))..).))))((((((...))))))......)))))).).))))))) ( -30.00) >DroYak_CAF1 3485 120 + 1 UAGUUUGGCACCGAAAGCGAAAUCCGAGUCCAGAAACCACUUGCCCUCUCAAUUCGGGCGAUCUUGAACCAACAAUUUCCGAUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....((((((((....)..........((((((((.....((((((.........)))))).....................(((((((...)))))))....)))))).)).))))))) ( -29.20) >consensus UAGUUUGGCACCGAAAGCCAAACCAGAGUCCAGAAACCACUUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAA ....(((((((.(...((((((...(((..(((.......)))..)))....((((((....))))))..............(((((((...))))))).....)))))).).))))))) (-28.52 = -28.40 + -0.12)

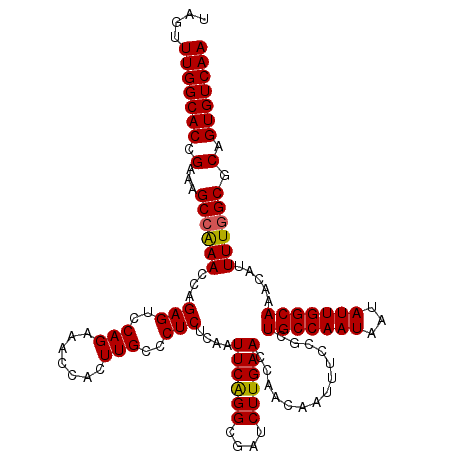

| Location | 20,272,054 – 20,272,174 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20272054 120 + 22407834 UUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCGCUGCUCGCUGCUCG ................(((((.(.(((((.............(((((((...)))))))........((((.((((((((.....)))....))))))))))))))..).)))))..... ( -27.70) >DroSec_CAF1 22927 108 + 1 UUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCGCU------------ (((((...........)))))....((((.............(((((((...)))))))........((((.((((((((.....)))....)))))))))))))...------------ ( -24.30) >DroSim_CAF1 21808 108 + 1 UUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCGCU------------ (((((...........)))))....((((.............(((((((...)))))))........((((.((((((((.....)))....)))))))))))))...------------ ( -24.30) >DroEre_CAF1 3114 119 + 1 UUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCGCUGCUCGCUGUUC- ................(((((.(.(((((.............(((((((...)))))))........((((.((((((((.....)))....))))))))))))))..).)))))....- ( -27.70) >DroYak_CAF1 3525 120 + 1 UUGCCCUCUCAAUUCGGGCGAUCUUGAACCAACAAUUUCCGAUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCCCUGUUCGCUGCUCG ((((((.........))))))....((((.(((.........(((((((...)))))))........((((.((((((((.....)))....))))))))))))....))))........ ( -28.40) >consensus UUGCCCUCUCAAUUCAGGCGAUCUUGAACCAACAAUUUCCGCUGCCAAUAAUAUUGGCAAACAUUUUGGCGCAGUGUCAAUUAAUUUGCCACACACUCGCUGUUCGCUG_UCGCUG_UC_ (((((...........)))))....((((.............(((((((...)))))))........((((.((((((((.....)))....)))))))))))))............... (-24.54 = -24.54 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:08 2006