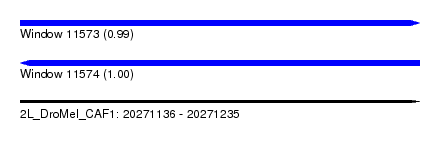
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,271,136 – 20,271,235 |
| Length | 99 |
| Max. P | 0.995819 |

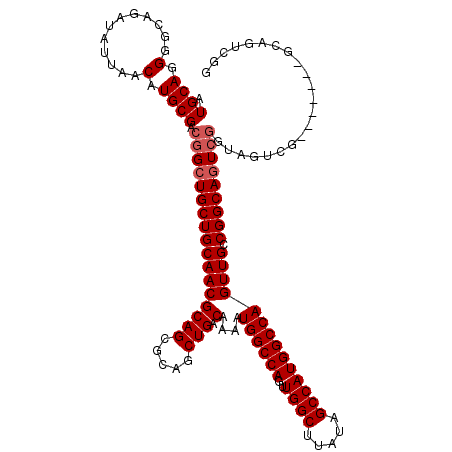
| Location | 20,271,136 – 20,271,235 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271136 99 + 22407834 CCG---------------------ACUGCCGGCAACUGGCCAUGGCUAUAAGCCAACUGGCCAUUUUGUCAGCUGCGCUGCGUUGCAGCAGCCGUCGCAUGUUAAUAUCUGCCCCUGCAU .((---------------------((.((.(((((.((((((((((.....))))..))))))..))))).((((((......)))))).)).))))............(((....))). ( -37.70) >DroSec_CAF1 21980 113 + 1 CCGACUAC-------CGACUGCCGACUGCCGGCAACUGGCCAUGGCUAUAAGCCAACUGGCCAUUUUGUCAGAUGCGCUGCGUUGCAGCAGCCGUCGCAUGUUAAUAUCUGCCCCUGCAU ........-------.((((((((.....)))))..((((((((((.....))))..))))))....))).((((.(((((......)))))))))(((.((........))...))).. ( -42.40) >DroSim_CAF1 20845 113 + 1 CCGACUGC-------CGACUACCGACUGCCGGCAACUGGCCAUGGCUAUAAGCCAACUGGCCAUUUUGUCAGAUGCGCUGCGUUGCAGCAGCCGUCGCAUGUUAAUAUCUGCCCCUGCAU ..((((((-------((.(........).)))))..((((((((((.....))))..))))))....))).((((.(((((......)))))))))(((.((........))...))).. ( -42.50) >DroEre_CAF1 2215 120 + 1 CCGACUGCCAACUGCCGACUACCGAAUGCCGGCAACUGGCCAUGGCUAUAAGCCAACUGGCCAUUUUGUCAGCUGCGCUGCGUUGCAGCAGCCGUCGCACGUUAAUAUCUGCCCCUGCAU ..(((.......(((((.(........).)))))..((((((((((.....))))..))))))....))).((((((......)))))).((.(..(((.((....)).)))..).)).. ( -40.40) >DroYak_CAF1 2207 113 + 1 CCGACUGC-------CGACUACCGACUGCCGGCAACUGGCCAUGGCCAUAAGCCAACUGGCCAUUUUGUCAGCUGCGCUGCGUUGCAGCAGCCGUCGCAUGUUAAUAUCUGCCCCUGCAU ..((((((-------.(((....(...((.(((((.((((((((((.....))))..))))))..))))).))..)(((((......))))).)))))).)))......(((....))). ( -40.10) >consensus CCGACUGC_______CGACUACCGACUGCCGGCAACUGGCCAUGGCUAUAAGCCAACUGGCCAUUUUGUCAGCUGCGCUGCGUUGCAGCAGCCGUCGCAUGUUAAUAUCUGCCCCUGCAU ..........................(((.((((..((((((((((.....))))..))))))........((.(((((((......)))).))).))...........))))...))). (-34.10 = -34.50 + 0.40)

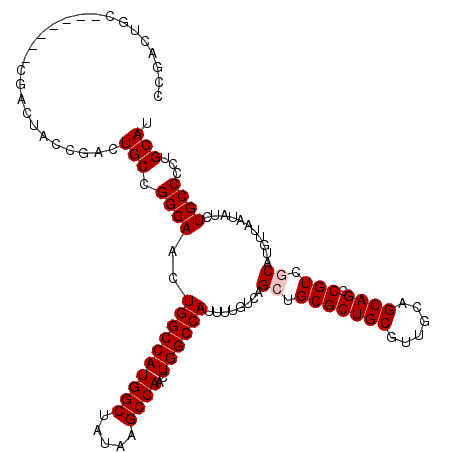
| Location | 20,271,136 – 20,271,235 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -47.84 |
| Consensus MFE | -38.92 |
| Energy contribution | -39.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20271136 99 - 22407834 AUGCAGGGGCAGAUAUUAACAUGCGACGGCUGCUGCAACGCAGCGCAGCUGACAAAAUGGCCAGUUGGCUUAUAGCCAUGGCCAGUUGCCGGCAGU---------------------CGG .((((.(............).)))).((((((((((((((((((...)))).)....((((((..((((.....)))))))))))))).)))))))---------------------)). ( -42.90) >DroSec_CAF1 21980 113 - 1 AUGCAGGGGCAGAUAUUAACAUGCGACGGCUGCUGCAACGCAGCGCAUCUGACAAAAUGGCCAGUUGGCUUAUAGCCAUGGCCAGUUGCCGGCAGUCGGCAGUCG-------GUAGUCGG ((((.(..(((..........)))..).(((((......)))))))))(((((....((((((..((((.....))))))))))..(((((((........))))-------)))))))) ( -46.20) >DroSim_CAF1 20845 113 - 1 AUGCAGGGGCAGAUAUUAACAUGCGACGGCUGCUGCAACGCAGCGCAUCUGACAAAAUGGCCAGUUGGCUUAUAGCCAUGGCCAGUUGCCGGCAGUCGGUAGUCG-------GCAGUCGG ((((.(..(((..........)))..).(((((......)))))))))(((((....((((((..((((.....))))))))))..(((((((........))))-------)))))))) ( -48.20) >DroEre_CAF1 2215 120 - 1 AUGCAGGGGCAGAUAUUAACGUGCGACGGCUGCUGCAACGCAGCGCAGCUGACAAAAUGGCCAGUUGGCUUAUAGCCAUGGCCAGUUGCCGGCAUUCGGUAGUCGGCAGUUGGCAGUCGG .((((.(............).)))).((((((((((...)))))...((..((....((((((..((((.....))))))))))..(((((((........)))))))))..))))))). ( -53.20) >DroYak_CAF1 2207 113 - 1 AUGCAGGGGCAGAUAUUAACAUGCGACGGCUGCUGCAACGCAGCGCAGCUGACAAAAUGGCCAGUUGGCUUAUGGCCAUGGCCAGUUGCCGGCAGUCGGUAGUCG-------GCAGUCGG .(((....)))............((((.((.(((((...))))))).((((((....((((((..((((.....))))))))))..(((((.....)))))))))-------)).)))). ( -48.70) >consensus AUGCAGGGGCAGAUAUUAACAUGCGACGGCUGCUGCAACGCAGCGCAGCUGACAAAAUGGCCAGUUGGCUUAUAGCCAUGGCCAGUUGCCGGCAGUCGGUAGUCG_______GCAGUCGG .((((.(............).)))).(((((((((((((((((.....))).)....((((((..((((.....)))))))))))))).)))))))))...................... (-38.92 = -39.52 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:03 2006