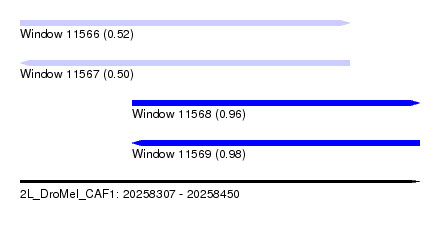
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,258,307 – 20,258,450 |
| Length | 143 |
| Max. P | 0.978713 |

| Location | 20,258,307 – 20,258,425 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20258307 118 + 22407834 CCAUGGGAACGCGGCGAAAAAAAAAUUGAUUUCGCAGAAAAAAUUCAAGCGCUGGAAGCAG--CAAAAAAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCG ..........((.((((((..........))))))........(((((((((((....)))--)..(((((((((....))))))))).....)))))))))..((((((....)))))) ( -34.20) >DroSec_CAF1 19860 115 + 1 CCAUGGGAACGCGGCGAAAA--AAAUUGAUUUCGCAGAAAAAAUGUUAGCGAUGGAAGCAG--CAAA-AAAGGGUGUAAAUCCUUUUUAUAAAAUUUGAAGCUACGAUAAUAAUUUAUCG ((((.(.((((..((((((.--.......))))))........))))..).)))).(((..--((((-(((((((....)))))))........))))..))).((((((....)))))) ( -24.90) >DroSim_CAF1 18715 116 + 1 CCAUGGGAACGCGGCGAAAA-AAAAUUGAUUUCGCGGAAAAAAUUCAAGCGAUGGAAGCAG--CAAA-AAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCG ..........((.((((((.-........))))))........((((((((.((....)).--).((-(((((((....))))))))).....)))))))))..((((((....)))))) ( -27.70) >DroEre_CAF1 87 113 + 1 CCAAAGAAAGACUGCGAAA---AAAUUGAUUUCGCUGAAAAAACGAAGGG---GAUGGCAGCGCAAA-AAAGGGUGUAAGUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAUUUUAUCG ....((..(((((((((((---.......)))))).........((((((---(((....((((...-.....))))..)))))))))....)))))....)).((((((....)))))) ( -24.80) >DroYak_CAF1 99 113 + 1 CCAAAGGAACGCUGCGAAA---AAAUUGAUUUCGCAGAAAAAAUAAAAGG-AUGAAGGCAG--CAAA-AAAGGGUGUAAAUCCGUUUUAUAAAGUUUGAAGCUGCGAUAAUAAUUUAUCG ...........((((((((---.......)))))))).............-......((((--(...-...((((....))))..((((.......)))))))))(((((....))))). ( -25.90) >consensus CCAUGGGAACGCGGCGAAAA__AAAUUGAUUUCGCAGAAAAAAUGAAAGCGAUGGAAGCAG__CAAA_AAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCG .............((((((..........)))))).....................(((....((((.(((((((....)))))))........))))..))).((((((....)))))) (-15.80 = -15.60 + -0.20)



| Location | 20,258,307 – 20,258,425 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20258307 118 - 22407834 CGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUUUUUUG--CUGCUUCCAGCGCUUGAAUUUUUUCUGCGAAAUCAAUUUUUUUUUCGCCGCGUUCCCAUGG .(((((....)))))(((((............(((((((........)))))))..)--))))....(((((..(((....))).((((((..........)))))).)))))....... ( -23.74) >DroSec_CAF1 19860 115 - 1 CGAUAAAUUAUUAUCGUAGCUUCAAAUUUUAUAAAAAGGAUUUACACCCUUU-UUUG--CUGCUUCCAUCGCUAACAUUUUUUCUGCGAAAUCAAUUU--UUUUCGCCGCGUUCCCAUGG .(((((....)))))(((((............(((((((........)))))-)).)--))))..(((((((.............((((((.......--.)))))).))).....)))) ( -20.26) >DroSim_CAF1 18715 116 - 1 CGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUU-UUUG--CUGCUUCCAUCGCUUGAAUUUUUUCCGCGAAAUCAAUUUU-UUUUCGCCGCGUUCCCAUGG .(((((....)))))(((((............(((((((........)))))-)).)--))))..(((((((..(((....))).((((((........-.)))))).))).....)))) ( -22.92) >DroEre_CAF1 87 113 - 1 CGAUAAAAUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGACUUACACCCUUU-UUUGCGCUGCCAUC---CCCUUCGUUUUUUCAGCGAAAUCAAUUU---UUUCGCAGUCUUUCUUUGG .(((((....)))))(((((..((((........(((((........)))))-)))).)))))....---...............((((((.......---))))))............. ( -18.70) >DroYak_CAF1 99 113 - 1 CGAUAAAUUAUUAUCGCAGCUUCAAACUUUAUAAAACGGAUUUACACCCUUU-UUUG--CUGCCUUCAU-CCUUUUAUUUUUUCUGCGAAAUCAAUUU---UUUCGCAGCGUUCCUUUGG .(((((....)))))(((((............((((.((........)).))-)).)--))))......-.............((((((((.......---))))))))........... ( -20.12) >consensus CGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUU_UUUG__CUGCUUCCAUCGCUUGAAUUUUUUCUGCGAAAUCAAUUU__UUUUCGCCGCGUUCCCAUGG .(((((....)))))((((...((((........(((((........))))).))))..))))......................((((((..........))))))............. (-13.54 = -13.58 + 0.04)

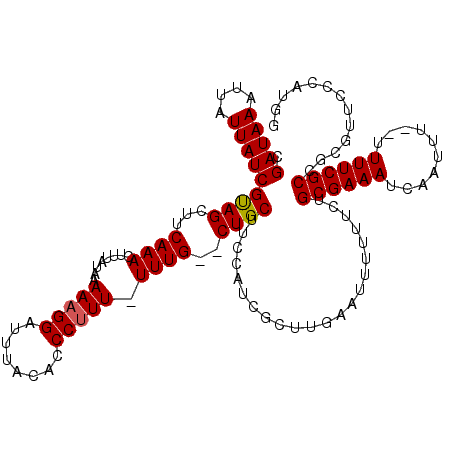
| Location | 20,258,347 – 20,258,450 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20258347 103 + 22407834 AAAUUCAAGCGCUGGAAGCAG--CAAAAAAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCGCAGCAUAAUGGGAGUUGAGACCGGC--------------- ...(((((((((((....)))--)..(((((((((....))))))))).....)))))))(((.((((((....)))))).)))....(((.........)))..--------------- ( -33.60) >DroSec_CAF1 19898 117 + 1 AAAUGUUAGCGAUGGAAGCAG--CAAA-AAAGGGUGUAAAUCCUUUUUAUAAAAUUUGAAGCUACGAUAAUAAUUUAUCGCAGCAUAAUGGUAGUUGAGAGCGGCAAAACCAUAAGCUGU .................((((--(.((-(((((((....)))))))))............(((.((((((....)))))).)))...(((((.((((....))))...)))))..))))) ( -30.80) >DroSim_CAF1 18754 117 + 1 AAAUUCAAGCGAUGGAAGCAG--CAAA-AAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCGCAGCAUAAUGGGAGUUGAGAGCGGCAAAACCAUAAGCUGU ...((((((((.((....)).--).((-(((((((....))))))))).....)))))))(((.((((((....)))))).)))...((((..((((....))))....))))....... ( -30.90) >DroEre_CAF1 124 115 + 1 AAACGAAGGG---GAUGGCAGCGCAAA-AAAGGGUGUAAGUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAUUUUAUCGCCGCAUAAUGGGAGUC-AGGGCGGGAAAACCAUAAGCUGU ....((((((---(((....((((...-.....))))..)))))))))...........((((.((((((....))))))((((....((.....)-)..))))..........)))).. ( -27.20) >DroYak_CAF1 136 116 + 1 AAAUAAAAGG-AUGAAGGCAG--CAAA-AAAGGGUGUAAAUCCGUUUUAUAAAGUUUGAAGCUGCGAUAAUAAUUUAUCGCAGCAUAAUGGGAGUCGAGAGCGGAAAAACCACAAGCUGU ..((((((((-((....(((.--(...-....).)))..)))).))))))..((((((..((((((((((....))))))))))....(((...(((....))).....))))))))).. ( -35.30) >consensus AAAUGAAAGCGAUGGAAGCAG__CAAA_AAAGGGUGUAAAUCCUUUUUAUAAAGUUUGAAGCUACGAUAAUAAUUUAUCGCAGCAUAAUGGGAGUUGAGAGCGGCAAAACCAUAAGCUGU ................(((.........(((((((....)))))))..............(((.((((((....)))))).))).........)))........................ (-15.46 = -15.22 + -0.24)

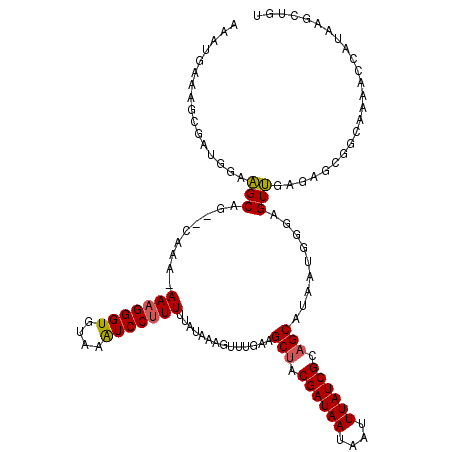
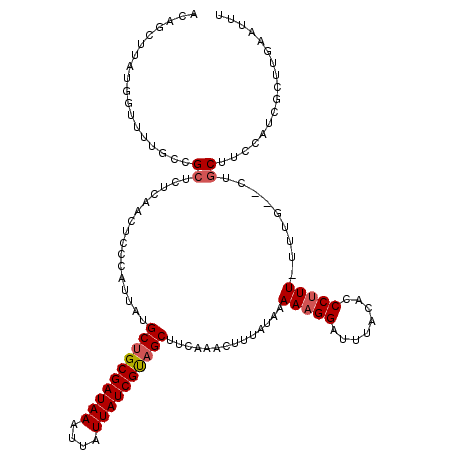
| Location | 20,258,347 – 20,258,450 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20258347 103 - 22407834 ---------------GCCGGUCUCAACUCCCAUUAUGCUGCGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUUUUUUG--CUGCUUCCAGCGCUUGAAUUU ---------------...((........))......((((((((((....))))))))))(((((.(.....(((((((........)))))))..(--(((....))))).)))))... ( -24.70) >DroSec_CAF1 19898 117 - 1 ACAGCUUAUGGUUUUGCCGCUCUCAACUACCAUUAUGCUGCGAUAAAUUAUUAUCGUAGCUUCAAAUUUUAUAAAAAGGAUUUACACCCUUU-UUUG--CUGCUUCCAUCGCUAACAUUU ..(((..((((....((.((................((((((((((....))))))))))............(((((((........)))))-)).)--).))..)))).)))....... ( -28.60) >DroSim_CAF1 18754 117 - 1 ACAGCUUAUGGUUUUGCCGCUCUCAACUCCCAUUAUGCUGCGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUU-UUUG--CUGCUUCCAUCGCUUGAAUUU ..(((..((((....((.((................((((((((((....))))))))))............(((((((........)))))-)).)--).))..)))).)))....... ( -28.00) >DroEre_CAF1 124 115 - 1 ACAGCUUAUGGUUUUCCCGCCCU-GACUCCCAUUAUGCGGCGAUAAAAUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGACUUACACCCUUU-UUUGCGCUGCCAUC---CCCUUCGUUU .((((....((.....))((...-............((.(((((((....))))))).))............(((((((........)))))-)).)))))).....---.......... ( -20.90) >DroYak_CAF1 136 116 - 1 ACAGCUUGUGGUUUUUCCGCUCUCGACUCCCAUUAUGCUGCGAUAAAUUAUUAUCGCAGCUUCAAACUUUAUAAAACGGAUUUACACCCUUU-UUUG--CUGCCUUCAU-CCUUUUAUUU .((((..((((.....))))................((((((((((....)))))))))).................((........))...-...)--))).......-.......... ( -24.70) >consensus ACAGCUUAUGGUUUUGCCGCUCUCAACUCCCAUUAUGCUGCGAUAAAUUAUUAUCGUAGCUUCAAACUUUAUAAAAAGGAUUUACACCCUUU_UUUG__CUGCUUCCAUCGCUUGAAUUU ..................((................((((((((((....))))))))))..............(((((........))))).........))................. (-14.90 = -15.34 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:58 2006