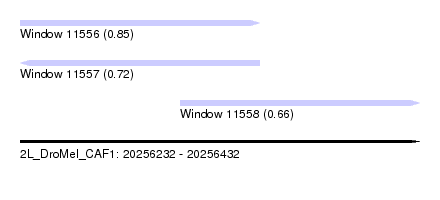
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,256,232 – 20,256,432 |
| Length | 200 |
| Max. P | 0.854859 |

| Location | 20,256,232 – 20,256,352 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -27.11 |
| Energy contribution | -26.34 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20256232 120 + 22407834 UGCAUUUUGCCCAUUAACACCUGUACAACACAUGCGAAACAUUAUUACCACCCGCGGCGAUAGGUAACUAUUUUAUUUAUUUUCCCCUCAUAUGCUUUGUUUCGCAAGGGCGUUAUUUUA ........((((.........(((.....)))(((((((((............(.((.(((((((((.....)))))))))..)))...........))))))))).))))......... ( -24.10) >DroSec_CAF1 17856 120 + 1 UGCAUUUCGCCCAUUAACACCUGCACAGCACAUGCGAAACAUUAUUACCACCCGCGGCGAUAGGGAACUAUUUUAUUUAUUUUCUCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUA .......(((((.........(((...)))..(((((((((.((((.((......)).(((((....)))))..................))))...))))))))).)))))........ ( -27.50) >DroSim_CAF1 16288 120 + 1 UGCAUUUCGCCCAUUAACACCUGCACAGCACAUGCGAAACAUUAUUACCACCCGCGGCGAUGGGGAACUAUUUUAUUUAUUUUCCCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUA .......(((((.........(((...)))..(((((((((.((((.((......)).((.((((((..............)))))))).))))...))))))))).)))))........ ( -32.74) >consensus UGCAUUUCGCCCAUUAACACCUGCACAGCACAUGCGAAACAUUAUUACCACCCGCGGCGAUAGGGAACUAUUUUAUUUAUUUUCCCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUA .......(((((.........(((...)))..(((((((((.(((..((......))..)))(((((..............)))))...........))))))))).)))))........ (-27.11 = -26.34 + -0.77)

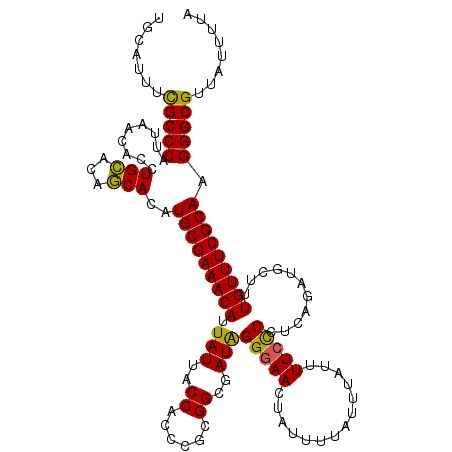

| Location | 20,256,232 – 20,256,352 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -29.84 |
| Energy contribution | -30.17 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723263 |
| Prediction | RNA |
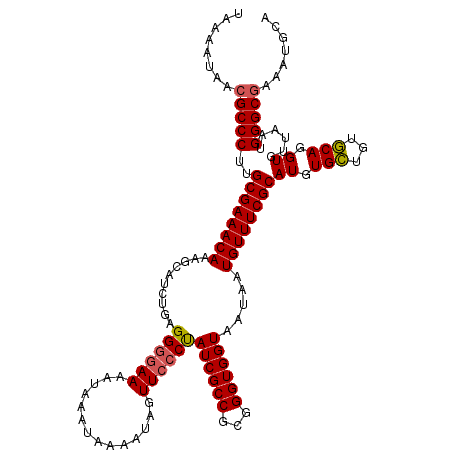
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20256232 120 - 22407834 UAAAAUAACGCCCUUGCGAAACAAAGCAUAUGAGGGGAAAAUAAAUAAAAUAGUUACCUAUCGCCGCGGGUGGUAAUAAUGUUUCGCAUGUGUUGUACAGGUGUUAAUGGGCAAAAUGCA .....(((((((..(((((((((..........((.....................))(((((((...)))))))....)))))))))((((...)))))))))))....((.....)). ( -28.20) >DroSec_CAF1 17856 120 - 1 UAAAAUAACGCCCUUGCGAAACAAAGCAUCUGAGGAGAAAAUAAAUAAAAUAGUUCCCUAUCGCCGCGGGUGGUAAUAAUGUUUCGCAUGUGCUGUGCAGGUGUUAAUGGGCGAAAUGCA ........(((((..((((((((.(.((((((.((.((.....(((......))).....)).)).)))))).).....))))))))((.(((...))).))......)))))....... ( -32.20) >DroSim_CAF1 16288 120 - 1 UAAAAUAACGCCCUUGCGAAACAAAGCAUCUGAGGGGAAAAUAAAUAAAAUAGUUCCCCAUCGCCGCGGGUGGUAAUAAUGUUUCGCAUGUGCUGUGCAGGUGUUAAUGGGCGAAAUGCA ........(((((..((((((((..........((((((..............))))))((((((...)))))).....))))))))((.(((...))).))......)))))....... ( -38.14) >consensus UAAAAUAACGCCCUUGCGAAACAAAGCAUCUGAGGGGAAAAUAAAUAAAAUAGUUCCCUAUCGCCGCGGGUGGUAAUAAUGUUUCGCAUGUGCUGUGCAGGUGUUAAUGGGCGAAAUGCA ........(((((..((((((((..........((((((..............))))))((((((...)))))).....))))))))((.(((...))).))......)))))....... (-29.84 = -30.17 + 0.34)


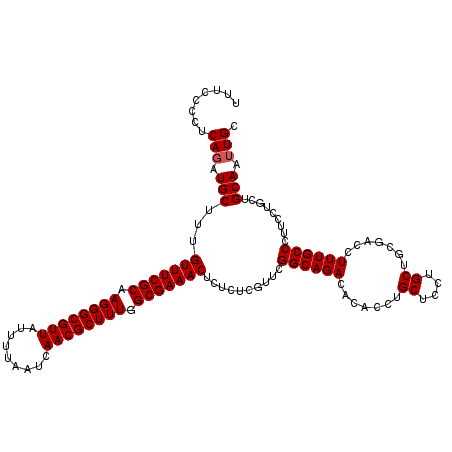
| Location | 20,256,312 – 20,256,432 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -30.11 |
| Energy contribution | -30.44 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20256312 120 + 22407834 UUUCCCCUCAUAUGCUUUGUUUCGCAAGGGCGUUAUUUUAAUCAACGCUUUGGCGAAACUCUCUCGUUCGGCAGACACACCUGCUCCUGCUGCGACCUUUGCCCUUCCUGCUGCAAUUGC ............(((...(((((((.((((((((.........)))))))).)))))))..........((((((.......((....)).......)))))).........)))..... ( -29.74) >DroSec_CAF1 17936 120 + 1 UUUCUCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUAAUCAACGCUUUGGCGAAACUCUCUCGUUCGGCAGACACACCUGCUCCUGCUGCGACCUUUGCCCUUCCUGCUGCAAUUGC ........(((.(((...(((((((.((((((((.........)))))))).)))))))..........((((((.......((....)).......)))))).........))).))). ( -30.64) >DroSim_CAF1 16368 120 + 1 UUUCCCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUAAUCAACGCUUUGGCGAAACUCUCUCGUUCGGCAGACACACCUGCUCCUGCUGCGACCUUUGCCCUUCCUGCUGCAAUUGC ........(((.(((...(((((((.((((((((.........)))))))).)))))))..........((((((.......((....)).......)))))).........))).))). ( -30.64) >consensus UUUCCCCUCAGAUGCUUUGUUUCGCAAGGGCGUUAUUUUAAUCAACGCUUUGGCGAAACUCUCUCGUUCGGCAGACACACCUGCUCCUGCUGCGACCUUUGCCCUUCCUGCUGCAAUUGC ........(((.(((...(((((((.((((((((.........)))))))).)))))))..........((((((.......((....)).......)))))).........))).))). (-30.11 = -30.44 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:48 2006