
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,255,229 – 20,255,428 |
| Length | 199 |
| Max. P | 0.991921 |

| Location | 20,255,229 – 20,255,349 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -38.99 |
| Energy contribution | -38.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20255229 120 + 22407834 AGGAUUGCAUAAGCUAAAGCCACCUGGCCAUAUUUCAGUGGUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCAUACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAU .((...((....)).....))((((((((((......)))).(((((.(.((((.(((...((.(((.((((....))))))))).)))...)).)).)))))))))))).......... ( -37.50) >DroSec_CAF1 16837 120 + 1 AGGAUUGCAUAAGCUAAAGCCACCUGGCCAUAUUUCAGUGGUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAU .((...((....)).....))((((((((((......)))).(((((.(.((((.(((...((.(((.((((....))))))))).)))...)).)).)))))))))))).......... ( -39.40) >DroSim_CAF1 15261 120 + 1 AGGAUUGCAUAAGCUAAAGCCACCUGGCCAUAUUUCAGUGGUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAU .((...((....)).....))((((((((((......)))).(((((.(.((((.(((...((.(((.((((....))))))))).)))...)).)).)))))))))))).......... ( -39.40) >consensus AGGAUUGCAUAAGCUAAAGCCACCUGGCCAUAUUUCAGUGGUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAU .((...((....)).....))((((((((((......)))).(((((.(.((((.(((...((.(((.((((....))))))))).)))...)).)).)))))))))))).......... (-38.99 = -38.77 + -0.22)

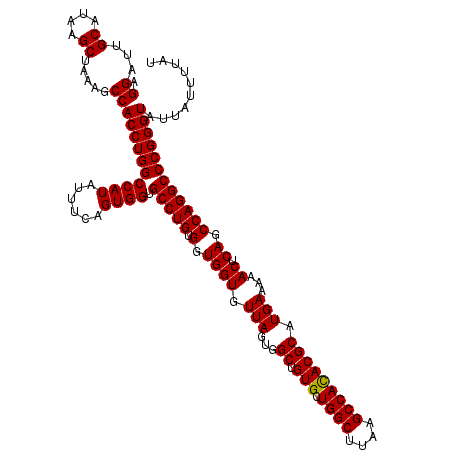
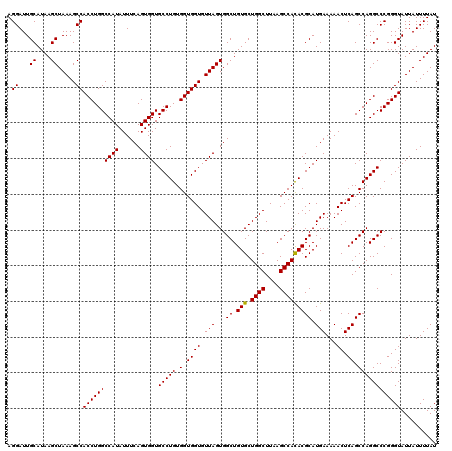
| Location | 20,255,269 – 20,255,389 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -45.99 |
| Energy contribution | -45.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20255269 120 + 22407834 GUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCAUACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGA ((((.((((((...((((((......))))))....)))))).))))......(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))). ( -44.50) >DroSec_CAF1 16877 120 + 1 GUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGA ((((.((((((...((((((......))))))....)))))).))))......(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))). ( -46.40) >DroSim_CAF1 15301 120 + 1 GUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGA ((((.((((((...((((((......))))))....)))))).))))......(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))). ( -46.40) >consensus GUGCCUGUGGUGGUGUUAGUGGCUGUGCUGGCUUAAGCCACACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGA ((((.((((((...((((((......))))))....)))))).))))......(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))). (-45.99 = -45.77 + -0.22)

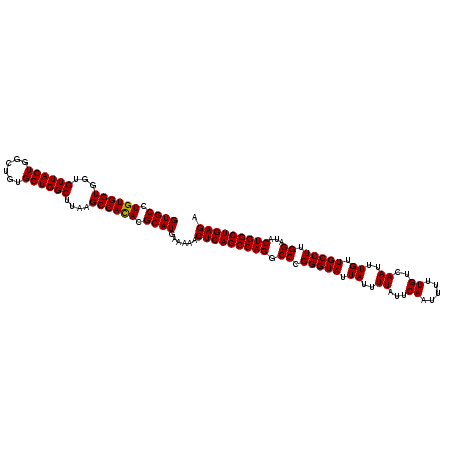

| Location | 20,255,269 – 20,255,389 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -30.79 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20255269 120 - 22407834 UCUCAGCCAGUAUCCAAGGCAACAAAUUGACAAAAAUUGAAUAAAAUAAUACCCGGGCCUGGCUGAGUUUUUCAUGCGUAUGGCUUAAGCCAGCACAGCCACUAACACCACCACAGGCAC .(((((((((..(((..(....).((((......))))................))).))))))))).......((.((.((((....)))))).))(((...............))).. ( -30.26) >DroSec_CAF1 16877 120 - 1 UCUCAGCCAGUAUCCAAGGCAACAAAUUGACAAAAAUUGAAUAAAAUAAUACCCGGGCCUGGCUGAGUUUUUCAUGCGUGUGGCUUAAGCCAGCACAGCCACUAACACCACCACAGGCAC .(((((((((..(((..(....).((((......))))................))).)))))))))..........(((((((....)))).))).(((...............))).. ( -32.56) >DroSim_CAF1 15301 120 - 1 UCUCAGCCAGUAUCCAAGGCAACAAAUUGACAAAAAUUGAAUAAAAUAAUACCCGGGCCUGGCUGAGUUUUUCAUGCGUGUGGCUUAAGCCAGCACAGCCACUAACACCACCACAGGCAC .(((((((((..(((..(....).((((......))))................))).)))))))))..........(((((((....)))).))).(((...............))).. ( -32.56) >consensus UCUCAGCCAGUAUCCAAGGCAACAAAUUGACAAAAAUUGAAUAAAAUAAUACCCGGGCCUGGCUGAGUUUUUCAUGCGUGUGGCUUAAGCCAGCACAGCCACUAACACCACCACAGGCAC .(((((((((..(((..(....).((((......))))................))).)))))))))..........(((((((....)))).))).(((...............))).. (-30.79 = -31.13 + 0.33)

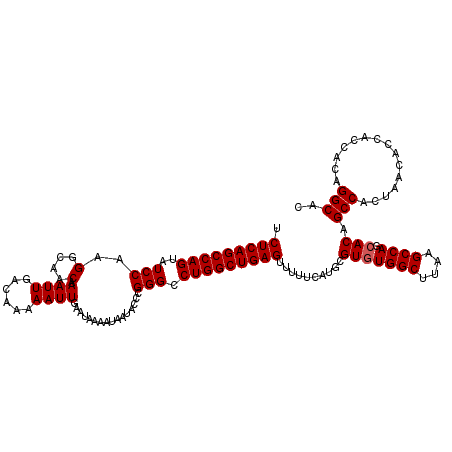
| Location | 20,255,309 – 20,255,428 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -38.42 |
| Energy contribution | -38.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20255309 119 + 22407834 UACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGAUCGGGGUGGGCAGC-UUGAGUGGGACUAAAACCAUUCAAU ..(((.(((....(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))).)))..)))......-((((((((........)))))))). ( -38.40) >DroSec_CAF1 16917 119 + 1 CACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGAUCGGGGUGGGCCCCGGUGAGUGGGAGUA-AACCAUUCAAU .............(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...)))))))))(((((((....)))))))((((((.....-..))))))... ( -46.30) >DroSim_CAF1 15341 118 + 1 CACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGAUCGGGGUGGGCCGC-UUGAGUGGGAGUA-AACCAUUCAAU ...((........(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))).....((....))))-((((((((.....-..)))))))). ( -39.30) >consensus CACGCAUGAAAAACUCAGCCAGGCCCGGGUAUUAUUUUAUUCAAUUUUUGUCAAUUUGUUGCCUUGGAUACUGGCUGAGAUCGGGGUGGGCCGC_UUGAGUGGGAGUA_AACCAUUCAAU (((.(.(((....(((((((((.((.(((((.((..((...((.....))..))..)).))))).))...))))))))).)))).)))........(((((((........))))))).. (-38.42 = -38.20 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:44 2006