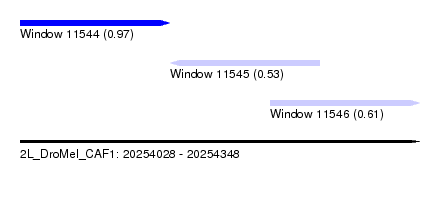
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,254,028 – 20,254,348 |
| Length | 320 |
| Max. P | 0.966862 |

| Location | 20,254,028 – 20,254,148 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -36.17 |
| Energy contribution | -37.67 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20254028 120 + 22407834 CGCUUCAUUCCUCUGCACAUUUCAUUGGUCAAUUGACUGUGAAAACCUGUGAAAACAAAAACACAUCCGUGGGAGGGACAGCAGAAGGACCGAGGUCCUGGCCAACAAGCCAUGGAAAAU ..((((..((((((((...((((((.((((....)))))))))).(((((....)).....(((....)))..)))....)))).))))..))))(((((((......)))).))).... ( -37.50) >DroSec_CAF1 15575 120 + 1 CGCUUCAUUCCUCUGCACAUUUCAUUGGUCAAUUGACUGUGAAAACCUGUGAAAACAAAAACACAACCGUGGGAGGGACAGCAGAAGGACCGAGGUCCUGGCCAACAAGCCAUGGAAAAU ..((((..((((((((...((((((.((((....)))))))))).(((((....)).....(((....)))..)))....)))).))))..))))(((((((......)))).))).... ( -37.50) >DroSim_CAF1 13994 120 + 1 CGCUUCAUUCCUCUGCACAUUUCAUUGGUCAAUUGACUGUGAAAACCUGUGAAAACAAAAACACGACCGUGGGAGGGACAGCAGAAGGACCGAGGUCCUGGCCAACAAGCCAUGGAAAAU ..((((..((((((((...((((((.((((....)))))))))).(((((....)).....(((....)))..)))....)))).))))..))))(((((((......)))).))).... ( -38.00) >consensus CGCUUCAUUCCUCUGCACAUUUCAUUGGUCAAUUGACUGUGAAAACCUGUGAAAACAAAAACACAACCGUGGGAGGGACAGCAGAAGGACCGAGGUCCUGGCCAACAAGCCAUGGAAAAU ..((((..((((((((...((((((.((((....)))))))))).(((((....)).....(((....)))..)))....)))).))))..))))(((((((......)))).))).... (-36.17 = -37.67 + 1.50)

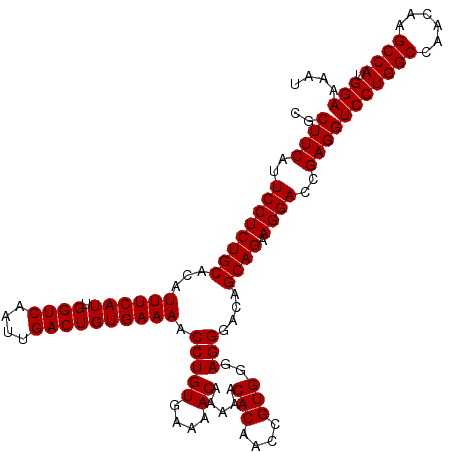
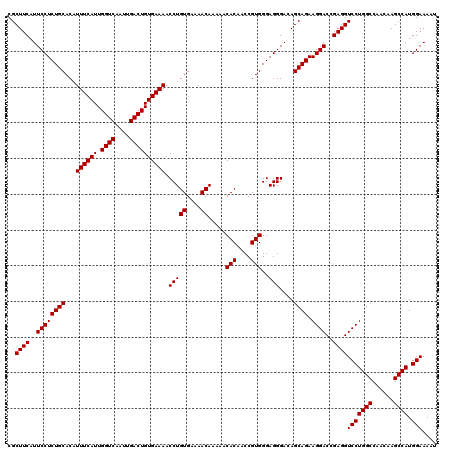
| Location | 20,254,148 – 20,254,268 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -32.21 |
| Energy contribution | -31.99 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.32 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20254148 120 - 22407834 UAAUGCUAAUGAUAUUUGCCUUAAAGAAUGGCAGCCGGAGUCGGGUUACAGGUAACAUCCAGUUAGCAGCUGCUGCCGGGGACUUUAGUUGAUGAAAAACUCUUUAGAAAUGCAUUAGUA ...((((((((.(((((...(((((((..((((((.(((.....((((....)))).)))(((.....)))))))))(....).................)))))))))))))))))))) ( -32.30) >DroSec_CAF1 15695 120 - 1 UAAUGCUAAUGAUAUUUGCCUUAAAGAAUGGCAGCCGGAGUCGGGUUACAGGUAACAUCCAGUUAGCAGCUGCUGCCGGGGACUUUAGUUGAUGAAAAACUCUUUAGAAAUGCAUUAGUA ...((((((((.(((((...(((((((..((((((.(((.....((((....)))).)))(((.....)))))))))(....).................)))))))))))))))))))) ( -32.30) >DroSim_CAF1 14114 120 - 1 UAAUGCUAAUGAUAUUUGCCUUAAAGAAUGGCAGCCGAAGUCGGGUUACAGGUAACAUCCAGUUAGCAGCUGCUGCCGGGGACUUUAGUUGAUGAAAAACUCUUUAGAAAUGCAUUAGUA ...((((((((.(((((...(((((((....((((..(((((..((((....)))).(((.((.(((....))))).))))))))..)))).........)))))))))))))))))))) ( -31.72) >consensus UAAUGCUAAUGAUAUUUGCCUUAAAGAAUGGCAGCCGGAGUCGGGUUACAGGUAACAUCCAGUUAGCAGCUGCUGCCGGGGACUUUAGUUGAUGAAAAACUCUUUAGAAAUGCAUUAGUA ...((((((((.(((((...(((((((....((((..(((((..((((....)))).(((.((.(((....))))).))))))))..)))).........)))))))))))))))))))) (-32.21 = -31.99 + -0.22)

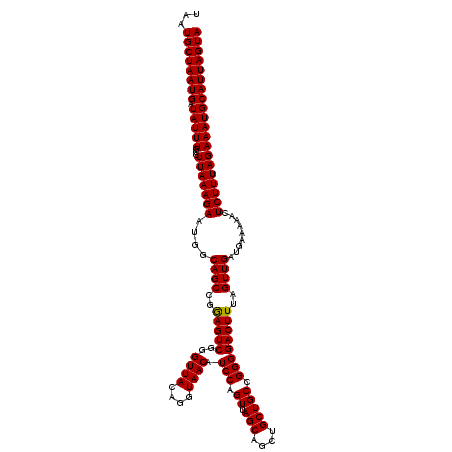

| Location | 20,254,228 – 20,254,348 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -28.13 |
| Energy contribution | -28.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20254228 120 + 22407834 CUCCGGCUGCCAUUCUUUAAGGCAAAUAUCAUUAGCAUUAUGAUGCCACUCCGCCCAACGACCACGCCACUUCCUCAGCCAUCCAGAUGGGUCCCAUCCUCGUCGCUCUUCGCAGUGGCC ....(((((((.........))))..((((((.......)))))).......)))..........((((((..............((((((......)).))))((.....)))))))). ( -28.60) >DroSec_CAF1 15775 120 + 1 CUCCGGCUGCCAUUCUUUAAGGCAAAUAUCAUUAGCAUUAUGAUGCCACUCCGCCCACCGACCACGCCACUUCCUCAGCCAUCCAGCUGGGUCCCAUCCUCGUCGCUCUUCGCAGUGGCC ....(((((((.........))))..((((((.......)))))).......)))..........((((((..((((((......)))))).............((.....)))))))). ( -30.30) >DroSim_CAF1 14194 120 + 1 CUUCGGCUGCCAUUCUUUAAGGCAAAUAUCAUUAGCAUUAUGAUGCCACUCCGCCCACCGACCACGCCACUUCCUCAGCCAUCCAGCUGGGUCCCAUCCUCGUCGCUCUUCGCAGUGGCC ....(((((((.........))))..((((((.......)))))).......)))..........((((((..((((((......)))))).............((.....)))))))). ( -30.30) >consensus CUCCGGCUGCCAUUCUUUAAGGCAAAUAUCAUUAGCAUUAUGAUGCCACUCCGCCCACCGACCACGCCACUUCCUCAGCCAUCCAGCUGGGUCCCAUCCUCGUCGCUCUUCGCAGUGGCC ....(((((((.........))))..((((((.......)))))).......)))..........((((((..((((((......)))))).............((.....)))))))). (-28.13 = -28.47 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:38 2006