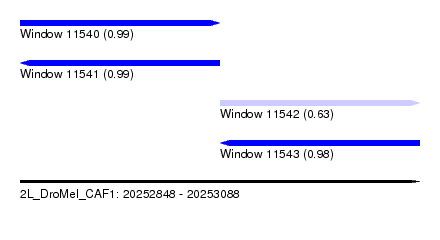
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,252,848 – 20,253,088 |
| Length | 240 |
| Max. P | 0.992737 |

| Location | 20,252,848 – 20,252,968 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -37.65 |
| Energy contribution | -37.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20252848 120 + 22407834 AGCGUGACAAUGUUUGUCGUCAAGUGUCGCAGACUCAUCGCUGAUAAUUGUUACGAUUAUCCACGCCCAUGCCAUCAUCAUGGCCUCGACGACAUCAUUAUCGCAGAACGCCCAGUUCCA ...((((.((((..(((((((..(((..((.((....)))).((((((((...)))))))))))......(((((....)))))...))))))).)))).)))).((((.....)))).. ( -35.60) >DroSec_CAF1 14369 117 + 1 AGCGUGACAAUGUUUGUCGUCAAGUGUCGCAGCCUCAUCGCUGAUAAUUGUUACGAUUAUCCACGCCCAUGCCAU---CAUGGCCACGACGACAUCAUUAUCACGGAACGCCCAGUUCCA ...((((.((((..(((((((..(((....(((......)))((((((((...)))))))))))(.(((((....---))))).)..))))))).)))).))))(((((.....))))). ( -41.30) >DroSim_CAF1 12784 117 + 1 AGCGUGACAAUGUUUGUCGUCAAGUGUCGCAGACUCAUCGCUGAUAAUUGUUACGAUUAUCCACGCCCAUGCCAU---CAUGGCCUCGACGACAUCAUUAUCACGGAACGCCCAGUUCCA ...((((.((((..(((((((..(((..((.((....)))).((((((((...)))))))))))(.(((((....---))))).)..))))))).)))).))))(((((.....))))). ( -39.70) >consensus AGCGUGACAAUGUUUGUCGUCAAGUGUCGCAGACUCAUCGCUGAUAAUUGUUACGAUUAUCCACGCCCAUGCCAU___CAUGGCCUCGACGACAUCAUUAUCACGGAACGCCCAGUUCCA ...((((.((((..(((((((..(((..((.........)).((((((((...)))))))))))......(((((....)))))...))))))).)))).))))(((((.....))))). (-37.65 = -37.77 + 0.11)

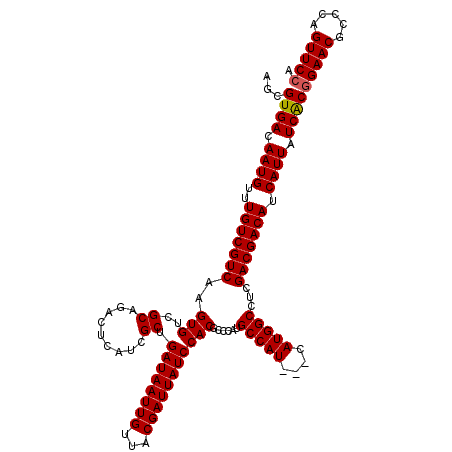
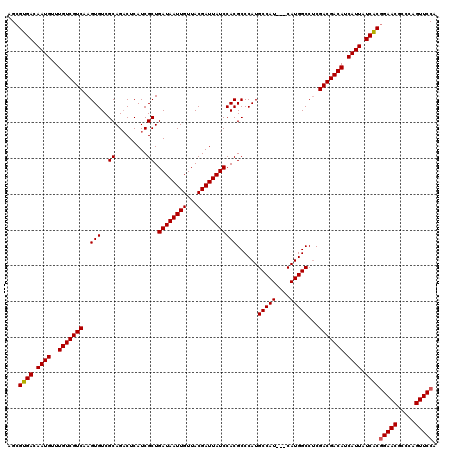
| Location | 20,252,848 – 20,252,968 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -47.87 |
| Consensus MFE | -47.09 |
| Energy contribution | -47.87 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20252848 120 - 22407834 UGGAACUGGGCGUUCUGCGAUAAUGAUGUCGUCGAGGCCAUGAUGAUGGCAUGGGCGUGGAUAAUCGUAACAAUUAUCAGCGAUGAGUCUGCGACACUUGACGACAAACAUUGUCACGCU .(((((.....)))))(.(((((((.(((((((((((((((....))))).((..((..(((.(((((...........)))))..)))..)).))))))))))))..))))))).)... ( -46.40) >DroSec_CAF1 14369 117 - 1 UGGAACUGGGCGUUCCGUGAUAAUGAUGUCGUCGUGGCCAUG---AUGGCAUGGGCGUGGAUAAUCGUAACAAUUAUCAGCGAUGAGGCUGCGACACUUGACGACAAACAUUGUCACGCU .(((((.....)))))(((((((((.((((((((((.(((((---....))))).((..(...(((((...........)))))....)..)).)))..)))))))..)))))))))... ( -45.70) >DroSim_CAF1 12784 117 - 1 UGGAACUGGGCGUUCCGUGAUAAUGAUGUCGUCGAGGCCAUG---AUGGCAUGGGCGUGGAUAAUCGUAACAAUUAUCAGCGAUGAGUCUGCGACACUUGACGACAAACAUUGUCACGCU .(((((.....)))))(((((((((.((((((((((.(((((---....))))).((..(((.(((((...........)))))..)))..))...))))))))))..)))))))))... ( -51.50) >consensus UGGAACUGGGCGUUCCGUGAUAAUGAUGUCGUCGAGGCCAUG___AUGGCAUGGGCGUGGAUAAUCGUAACAAUUAUCAGCGAUGAGUCUGCGACACUUGACGACAAACAUUGUCACGCU .(((((.....)))))(((((((((.(((((((((((((((....))))).((..((..(((.(((((...........)))))..)))..)).))))))))))))..)))))))))... (-47.09 = -47.87 + 0.78)

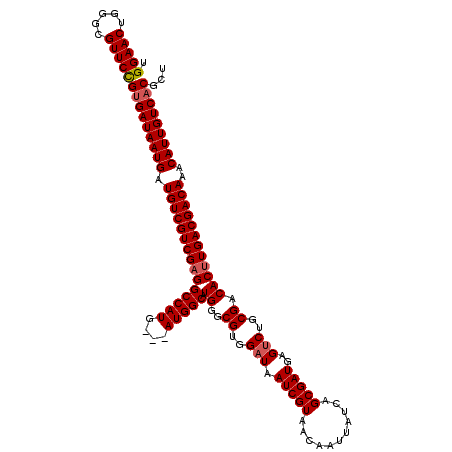
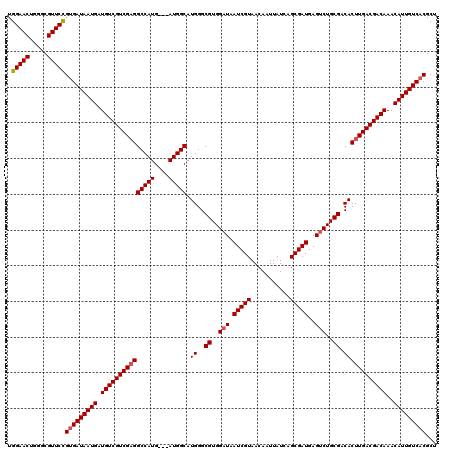
| Location | 20,252,968 – 20,253,088 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20252968 120 + 22407834 CUUCGAAUUUAGUGGGAGUUGUGUGUGAGGGGUAAAUCCAUUCGACCUAUUCCUUACACUAAUGAUUUAAUUAUGUUCCCGCUGCGAUUGUCGAGUGGGGAUCAAAUCACAUUGACAAAG ..(((....(((((((((....((((((((((((..((.....))..))))))))))))((((......))))..))))))))))))(((((((((((........)))).))))))).. ( -38.50) >DroSec_CAF1 14486 120 + 1 CUUCGAAUUUAGUGUGAGUUGUGUGCGAGGGGUAAAUCCAUUCGACCUAUUCCUUACACUCAUGAUUUAAUUAUGUUCCCGCUGCGAUUGUCGAGUGGGGAUCAAAUCACGAUGACGAAG (((((......(((((((..(((.(((((.((.....)).)))).).)))..)))))))((.((((((......(((((((((.((.....))))))))))).)))))).))...))))) ( -38.60) >DroSim_CAF1 12901 120 + 1 CUUCGAAUUUAGUGUGAGUUGUGUGCGAGGGGUAAAUCCAUUCGACCUAUUCCUUACACUCAUGAUUUAAUUAUGUUCCCGCUGCGAUUGUCGAGUGGGGAUCAAAUCACGAUGACAAAG ..(((.....((((((((..(((.(((((.((.....)).)))).).)))..))))))))..((((((......(((((((((.((.....))))))))))).)))))))))........ ( -35.20) >consensus CUUCGAAUUUAGUGUGAGUUGUGUGCGAGGGGUAAAUCCAUUCGACCUAUUCCUUACACUCAUGAUUUAAUUAUGUUCCCGCUGCGAUUGUCGAGUGGGGAUCAAAUCACGAUGACAAAG ..........((((((((..(((.(((((.((.....)).)))).).)))..))))))))..((((((......(((((((((.((.....))))))))))).))))))........... (-31.49 = -31.60 + 0.11)

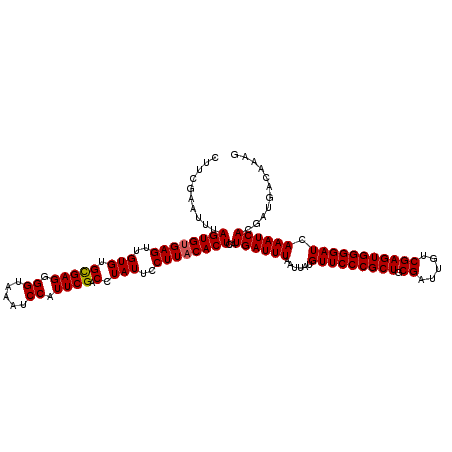
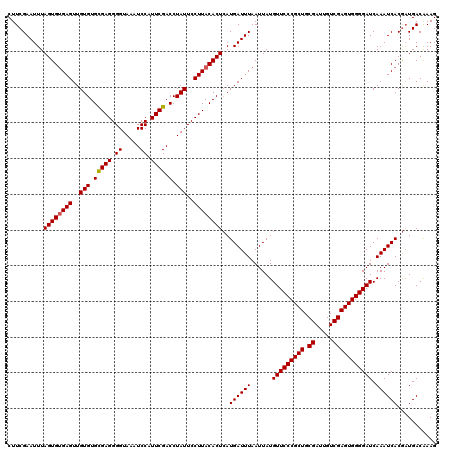
| Location | 20,252,968 – 20,253,088 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.57 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20252968 120 - 22407834 CUUUGUCAAUGUGAUUUGAUCCCCACUCGACAAUCGCAGCGGGAACAUAAUUAAAUCAUUAGUGUAAGGAAUAGGUCGAAUGGAUUUACCCCUCACACACAACUCCCACUAAAUUCGAAG (((((.....((((((((((.(((.((((.....)).)).)))......))))))))))..((((.(((..((((((.....))))))..))).)))).................))))) ( -27.70) >DroSec_CAF1 14486 120 - 1 CUUCGUCAUCGUGAUUUGAUCCCCACUCGACAAUCGCAGCGGGAACAUAAUUAAAUCAUGAGUGUAAGGAAUAGGUCGAAUGGAUUUACCCCUCGCACACAACUCACACUAAAUUCGAAG (((((...((((((((((((.(((.((((.....)).)).)))......))))))))))))(((..(((..((((((.....))))))..)))..))).................))))) ( -35.40) >DroSim_CAF1 12901 120 - 1 CUUUGUCAUCGUGAUUUGAUCCCCACUCGACAAUCGCAGCGGGAACAUAAUUAAAUCAUGAGUGUAAGGAAUAGGUCGAAUGGAUUUACCCCUCGCACACAACUCACACUAAAUUCGAAG (((((...((((((((((((.(((.((((.....)).)).)))......))))))))))))(((..(((..((((((.....))))))..)))..))).................))))) ( -33.30) >consensus CUUUGUCAUCGUGAUUUGAUCCCCACUCGACAAUCGCAGCGGGAACAUAAUUAAAUCAUGAGUGUAAGGAAUAGGUCGAAUGGAUUUACCCCUCGCACACAACUCACACUAAAUUCGAAG (((((...((((((((((((.(((.((((.....)).)).)))......))))))))))))((((.(((..((((((.....))))))..))).)))).................))))) (-31.34 = -31.57 + 0.23)

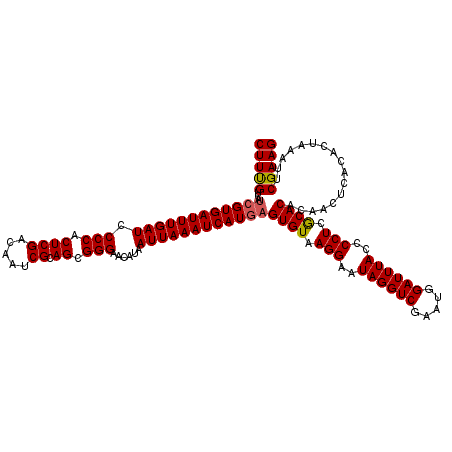
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:35 2006