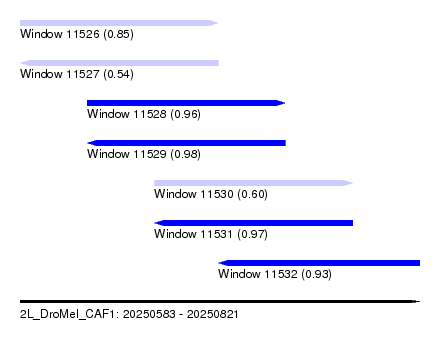
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,250,583 – 20,250,821 |
| Length | 238 |
| Max. P | 0.981203 |

| Location | 20,250,583 – 20,250,701 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -32.63 |
| Energy contribution | -32.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250583 118 + 22407834 AGAACGUGGCUAAACACCCAACUUGCGAUGCCCUUUGCUCAUUCGCCUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU-- ...((((((((((...........((((.((.....))....)))).(((((..(.(((((((((((......)))))))))))...)...))))).......))))))...))))..-- ( -28.80) >DroSec_CAF1 12169 120 + 1 AGAACGUGGCUAAACACCCAACUCGCGAUGGCCUUUGCUCAUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUCG ((((..(((((..((((.........(((((((..(((......))).))))))).(((((((((((......))))))))))).))))..)))))..)))).................. ( -36.20) >DroSim_CAF1 10578 120 + 1 AGAACGUGGCUAAACACCCAACUUGCGAUGGCCUUUGCUCAUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUAG ((((..(((((..((((.........(((((((..(((......))).))))))).(((((((((((......))))))))))).))))..)))))..)))).................. ( -36.20) >consensus AGAACGUGGCUAAACACCCAACUUGCGAUGGCCUUUGCUCAUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU_G ((((..(((((..((((.........(((((((...((......))..))))))).(((((((((((......))))))))))).))))..)))))..)))).................. (-32.63 = -32.97 + 0.33)


| Location | 20,250,583 – 20,250,701 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250583 118 - 22407834 --ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCAGGCGAAUGAGCAAAGGGCAUCGCAAGUUGGGUGUUUAGCCACGUUCU --((((.((((.((((((....)))))).((..............(((..(((....)))..)))((((.((..((......))...)).))))))..)))).))))............. ( -25.50) >DroSec_CAF1 12169 120 - 1 CGACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAUGAGCAAAGGCCAUCGCGAGUUGGGUGUUUAGCCACGUUCU .(((((.((((.((((((....))))))(((..............(((..(((....)))..)))(((((((..((......))...)))))))))).)))).)))))............ ( -32.40) >DroSim_CAF1 10578 120 - 1 CUACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAUGAGCAAAGGCCAUCGCAAGUUGGGUGUUUAGCCACGUUCU ..((((.((((.((((((....)))))).((..............(((..(((....)))..)))(((((((..((......))...)))))))))..)))).))))............. ( -29.20) >consensus C_ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAUGAGCAAAGGCCAUCGCAAGUUGGGUGUUUAGCCACGUUCU ..((((.((((.((((((....)))))).((..............(((..(((....)))..)))(((((((..((......))...)))))))))..)))).))))............. (-27.00 = -27.33 + 0.33)

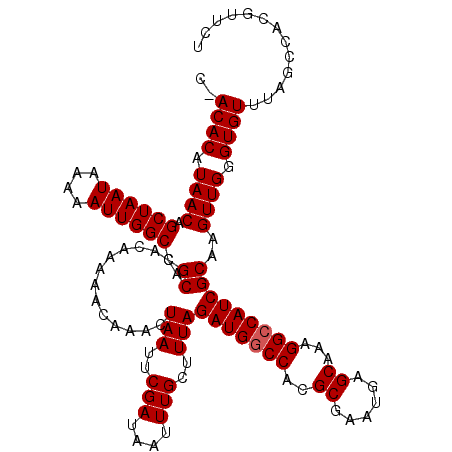
| Location | 20,250,623 – 20,250,741 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -35.15 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250623 118 + 22407834 AUUCGCCUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU--AGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCGUGCUCAA .(((.((((((((...(((((((((((......)))))))))))...(((((((.(((....))).((((.......)--)))))))))).))))))))))).................. ( -39.20) >DroSec_CAF1 12209 120 + 1 AUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUCGAGCGUUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAA ...(((.(((((((..(((((((((((......))))))))))).))).(((((.(((....))).))))).....)))).))).......(((.((((((((....)))))))).))). ( -36.90) >DroSim_CAF1 10618 120 + 1 AUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUAGAGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAG ....((..(((((...(((((((((((......)))))))))))...(((((((.(((....))).(((.(.......).)))))))))).)))))(((((((....))))))))).... ( -39.50) >consensus AUUCGCGUGGCCAUCUAAAGCAAAUUAUCGAAUUAGUUUGUUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU_GAGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAA ....((..(((((...(((((((((((......)))))))))))...(((((((.(((....))).(((...........)))))))))).)))))(((((((....))))))))).... (-35.15 = -35.27 + 0.11)

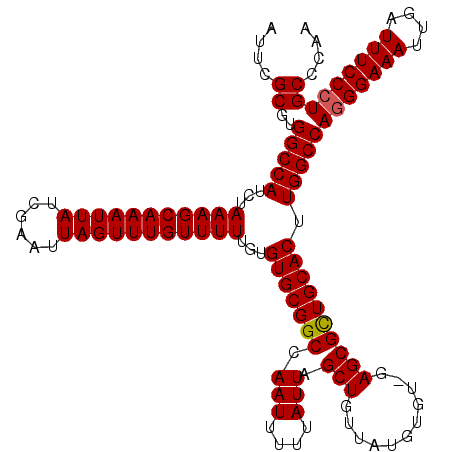
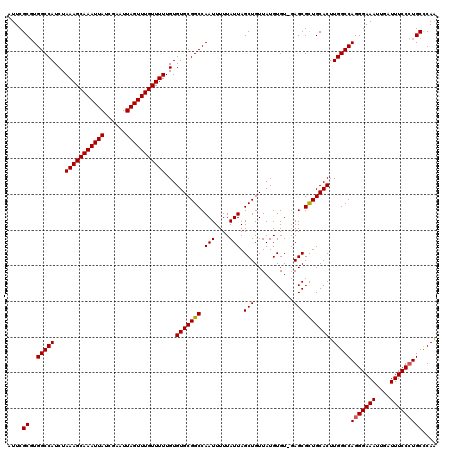
| Location | 20,250,623 – 20,250,741 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -30.43 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250623 118 - 22407834 UUGAGCACGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCU--ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCAGGCGAAU ....((..(((((....)))))(((((((.((((.(((((--.........)))((((....)))))).))))...........((((..(((....)))..)))).))))))))).... ( -31.80) >DroSec_CAF1 12209 120 - 1 UUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAACGCUCGACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAU ....(((((((((....)))))))(((((.((((..(....)..........((((((....)))))).))))...........((((..(((....)))..)))).)))))..)).... ( -33.10) >DroSim_CAF1 10618 120 - 1 CUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCUCUACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAU ....(((((((((....)))))))(((((.((((.(.(.....).)......((((((....)))))).))))...........((((..(((....)))..)))).)))))..)).... ( -32.90) >consensus UUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCUC_ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAACAAACUAAUUCGAUAAUUUGCUUUAGAUGGCCACGCGAAU ....(((((((((....)))))))(((((.((((...(.....)........((((((....)))))).))))...........((((..(((....)))..)))).)))))..)).... (-30.43 = -30.77 + 0.33)

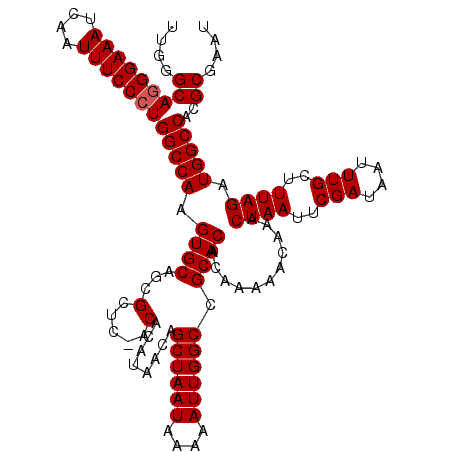

| Location | 20,250,663 – 20,250,781 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -31.27 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250663 118 + 22407834 UUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU--AGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCGUGCUCAAAGAAGCUGGCCAAAACAGAAUAAUUCCGGACAGUAAAUUA .(((((((((((((.(((....))).((((.......)--))))))))))(((((((.(((((....)))))..(((......)))))))))).)))))).................... ( -32.90) >DroSec_CAF1 12249 120 + 1 UUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUCGAGCGUUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAAAGAAGCUGGCCAAAACAAGAUAAUUCCGGACAGUAAAUUA .((((((....(((((.(((((......(((.((((.....)))).))).((((.((((((((....)))))))).))))))))).)))))...)))))).................... ( -34.30) >DroSim_CAF1 10658 120 + 1 UUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGUAGAGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAGAGAAGCUGGCCAAAACAAGAUAAUUCCGGACAGUAAAUUA .((((((....(((((.(((((............((((((....))))))((((.((((((((....)))))))).))))))))).)))))...)))))).................... ( -37.40) >consensus UUUUUGUGUGCGGCCAAUUUUUAUUAGCUGUUAUGUGU_GAGCGCUGCACUUGGCCAGGGAAAUUGAUUUCCCUGCCCAAAGAAGCUGGCCAAAACAAGAUAAUUCCGGACAGUAAAUUA .((((((....(((((.(((((..((((.(((........)))))))...((((.((((((((....)))))))).))))))))).)))))...)))))).................... (-31.27 = -30.50 + -0.77)



| Location | 20,250,663 – 20,250,781 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -30.83 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250663 118 - 22407834 UAAUUUACUGUCCGGAAUUAUUCUGUUUUGGCCAGCUUCUUUGAGCACGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCU--ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAA ........(((.((........((((((((((((((((....))))..(((((....))))).)))))))).))))....--..........((((((....)))))))).)))...... ( -28.90) >DroSec_CAF1 12249 120 - 1 UAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUUUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAACGCUCGACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAA .......(((.(((((((......))))))).))).....((((.((((((((....)))))))).))))((((..(....)..........((((((....)))))).))))....... ( -36.90) >DroSim_CAF1 10658 120 - 1 UAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUCUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCUCUACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAA .......(((.(((((((......))))))).)))......(((.((((((((....)))))))).))).((((.(.(.....).)......((((((....)))))).))))....... ( -35.70) >consensus UAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUUUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCUC_ACACAUAACAGCUAAUAAAAAUUGGCCGCACACAAAAA .......(((.(((((((......))))))).)))......(((.((((((((....)))))))).))).((((...(.....)........((((((....)))))).))))....... (-30.83 = -31.50 + 0.67)

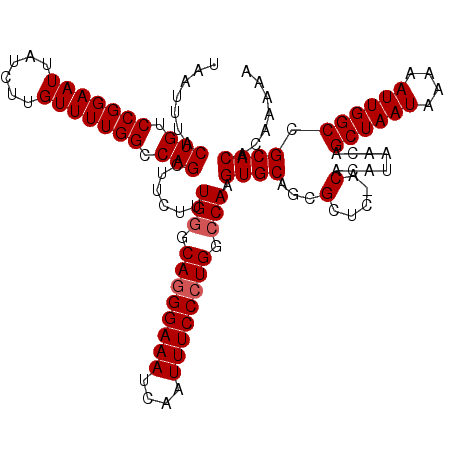
| Location | 20,250,701 – 20,250,821 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -43.64 |
| Consensus MFE | -41.26 |
| Energy contribution | -40.93 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20250701 120 - 22407834 CAAUUAGGGGGAUAGCCCGCAGCUGUCUGCGGGGCAAACUUAAUUUACUGUCCGGAAUUAUUCUGUUUUGGCCAGCUUCUUUGAGCACGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCU .....((.((((((((((((((....)))))))..............)))))).........((((((((((((((((....))))..(((((....))))).)))))))).))))).)) ( -40.33) >DroSec_CAF1 12289 120 - 1 CAAUUAGGGGGAUAGCCCGCAGCUGUCUGCGGGGCAAACUUAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUUUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAACGCU .........(((((((.....)))))))(((..(((...........(((.(((((((......))))))).)))....(((((.((((((((....)))))))).))))))))..))). ( -46.20) >DroSim_CAF1 10698 120 - 1 CAAUUAGGGGGAUAGCCCGCAGCUGUCUGCGGGGCAAACUUAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUCUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCU .(((((((..(....(((((((....))))))).)...)))))))...((.(((((((......))))))).))(((.(.((((.((((((((....)))))))).)).)).).)))... ( -44.40) >consensus CAAUUAGGGGGAUAGCCCGCAGCUGUCUGCGGGGCAAACUUAAUUUACUGUCCGGAAUUAUCUUGUUUUGGCCAGCUUCUUUGGGCAGGGAAAUCAAUUUCCCUGGCCAAGUGCAGCGCU .....((.((((((((((((((....)))))))..............)))))).........(((((((((((.(((......)))(((((((....)))))))))))))).))))).)) (-41.26 = -40.93 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:25 2006