
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,247,890 – 20,248,210 |
| Length | 320 |
| Max. P | 0.987386 |

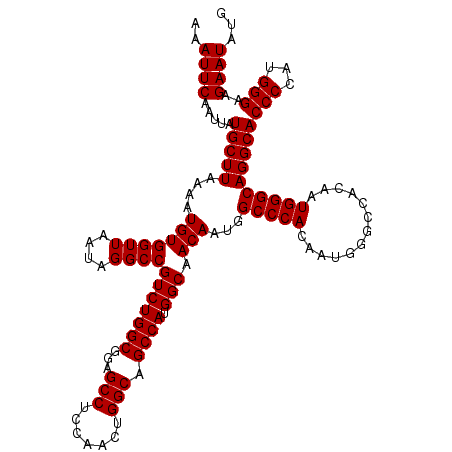
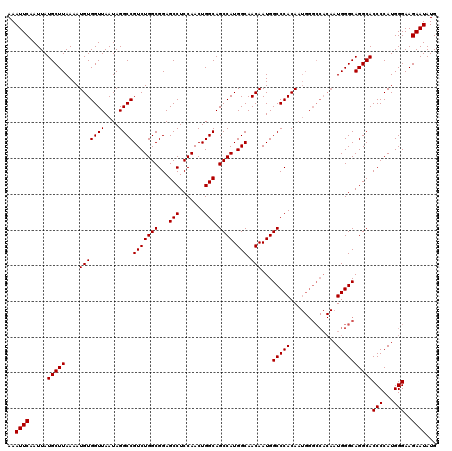
| Location | 20,247,890 – 20,248,010 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247890 120 - 22407834 AAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCACAAUGGGCCACAAUGGGCAGGCACCCCAUGGGAAGAAUAUG ..((((.....((((((...(((((((....))))(((((((...(((.......))).)))).))).))).((((((.....))))))...))))))....(((...)))..))))... ( -40.30) >DroSec_CAF1 6421 110 - 1 AAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCACA----------AUGGGCAGGCACCCCAUGGGAAGAAUAUG ..((((.....((((((...(((((.......((((..(((.(....).)))..)))).((((((....).))))))))))----------.))))))....(((...)))..))))... ( -35.00) >DroSim_CAF1 6863 120 - 1 AAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCACAAUGGGCCACAAUGGGCAGGCACCCCAUGGGAAGAAUAUG ..((((.....((((((...(((((((....))))(((((((...(((.......))).)))).))).))).((((((.....))))))...))))))....(((...)))..))))... ( -40.30) >consensus AAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCACAAUGGGCCACAAUGGGCAGGCACCCCAUGGGAAGAAUAUG ..((((.....(((((....(((((((....))))(((((((...(((.......))).)))).))).)))...(((((.............))))))))))(((...)))..))))... (-34.22 = -34.22 + -0.00)

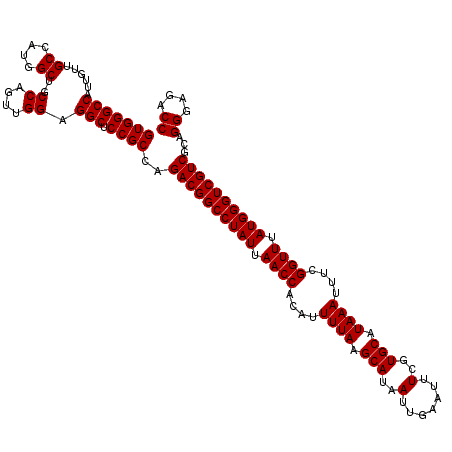
| Location | 20,247,930 – 20,248,050 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -43.00 |
| Energy contribution | -43.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247930 120 + 22407834 GUGGGCCAUUGUUGCCAUGGCUGCCAGUUGGAGGCUCCGCCAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACC (((((((......((....))..((....)).))).))))..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....)) ( -43.00) >DroSec_CAF1 6451 120 + 1 GUGGGCCAUUGUUGCCAUGGCUGCCAGUUGGAGGCUCCGCCAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACC (((((((......((....))..((....)).))).))))..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....)) ( -43.00) >DroSim_CAF1 6903 120 + 1 GUGGGCCAUUGUUGCCAUGGCUGCCAGUUGGAGGCUCCGCCAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACC (((((((......((....))..((....)).))).))))..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....)) ( -43.00) >consensus GUGGGCCAUUGUUGCCAUGGCUGCCAGUUGGAGGCUCCGCCAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACC (((((((......((....))..((....)).))).))))..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....)) (-43.00 = -43.00 + 0.00)


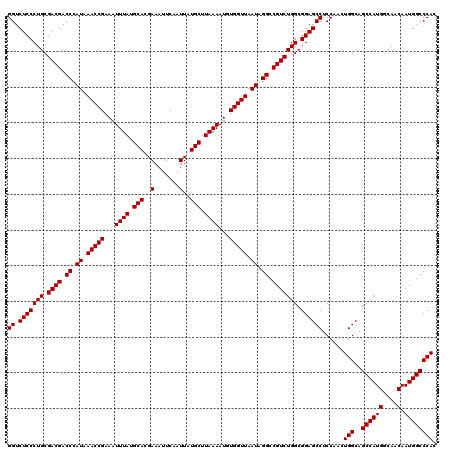
| Location | 20,247,930 – 20,248,050 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247930 120 - 22407834 GGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCAC ((.(((((((.((((.((.((.(((((...((((.(((..(.......)..))).))))...))))).)).)).))))))).))))))......(((..((((((....).)))))))). ( -37.60) >DroSec_CAF1 6451 120 - 1 GGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCAC ((.(((((((.((((.((.((.(((((...((((.(((..(.......)..))).))))...))))).)).)).))))))).))))))......(((..((((((....).)))))))). ( -37.60) >DroSim_CAF1 6903 120 - 1 GGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCAC ((.(((((((.((((.((.((.(((((...((((.(((..(.......)..))).))))...))))).)).)).))))))).))))))......(((..((((((....).)))))))). ( -37.60) >consensus GGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUGGCGGAGCCUCCAACUGGCAGCCAUGGCAACAAUGGCCCAC ((.(((((((.((((.((.((.(((((...((((.(((..(.......)..))).))))...))))).)).)).))))))).))))))......(((..((((((....).)))))))). (-37.60 = -37.60 + 0.00)


| Location | 20,247,970 – 20,248,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -38.72 |
| Energy contribution | -38.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247970 120 + 22407834 CAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACCACAACCCGGCCAGAUUAUUACAUUAUUUACUGGGUGUGUG ..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....))(((((((((..((((.........)))).)))))).))). ( -39.60) >DroSec_CAF1 6491 120 + 1 CAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACCACAACCCGGCCAGAUUAUUACAUUAUUUACUGGGCGUGUG ..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....))...((.((.((((...((......))...)))).)).)). ( -39.20) >DroSim_CAF1 6943 120 + 1 CAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACCACAACCCGGCCAGAUUAUUACAUUAUUUACUGGGCGUGUG ..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....))...((.((.((((...((......))...)))).)).)). ( -39.20) >consensus CAGACGGCCUAUUAACCACAUUUUAAGCAUAAUUGAAUUUCGUGCAUAAAUUUCGGUUUAUGGGUCGUCGCAGGGAGACCACAACCCGGCCAGAUUAUUACAUUAUUUACUGGGCGUGUG ..((((((((((.((((....((((.(((..(.......)..))).))))....)))).))))))))))...((....))...((.((.((((...((......))...)))).)).)). (-38.72 = -38.50 + -0.22)



| Location | 20,247,970 – 20,248,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247970 120 - 22407834 CACACACCCAGUAAAUAAUGUAAUAAUCUGGCCGGGUUGUGGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUG .............................(((((((((((.((......)).)))))))...(((((...((((.(((..(.......)..))).))))...)))))....))))..... ( -31.00) >DroSec_CAF1 6491 120 - 1 CACACGCCCAGUAAAUAAUGUAAUAAUCUGGCCGGGUUGUGGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUG .............................(((((((((((.((......)).)))))))...(((((...((((.(((..(.......)..))).))))...)))))....))))..... ( -31.00) >DroSim_CAF1 6943 120 - 1 CACACGCCCAGUAAAUAAUGUAAUAAUCUGGCCGGGUUGUGGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUG .............................(((((((((((.((......)).)))))))...(((((...((((.(((..(.......)..))).))))...)))))....))))..... ( -31.00) >consensus CACACGCCCAGUAAAUAAUGUAAUAAUCUGGCCGGGUUGUGGUCUCCCUGCGACGACCCAUAAACCGAAAUUUAUGCACGAAAUUCAAUUAUGCUUAAAAUGUGGUUAAUAGGCCGUCUG .............................(((((((((((.((......)).)))))))...(((((...((((.(((..(.......)..))).))))...)))))....))))..... (-31.00 = -31.00 + 0.00)

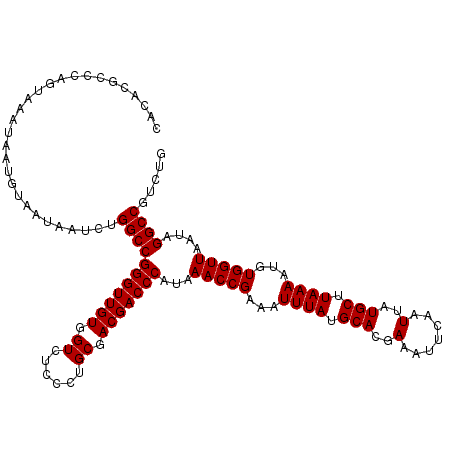
| Location | 20,248,090 – 20,248,210 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -42.94 |
| Energy contribution | -43.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20248090 120 - 22407834 AAACAACAGAGGGAGGCAGGACGAAACGUGGAAUCAUGAAAAAUUGAUGCCCCUGUCGUCUGGUGGCGCCACCUGGUCUGCCCUCUCUCUCGCACGGCAGCUGGCCGUCUCUUUCGUGCA .......(((((((((((((((((...(.((.((((........)))).)))...))))).((((....))))....))).))))))))).(((((((.....)))(.......))))). ( -45.60) >DroSec_CAF1 6611 120 - 1 AAACAACAGAGGGAAGCAGGCCGAAACGUGGAAUCAUGAAAAAUUGAUGCCCCUGUCGUCUGGUGGCGCCACCUGGUCUGCCCUCUCUCUCGCACGGCAUCUGGCCGUCUCUUUCGUGCA .......(((((((.((((((((....((((.((((........))))(((((........)).))).)))).))))))))..))))))).(((((((.....)))(.......))))). ( -45.00) >DroSim_CAF1 7063 120 - 1 AAACAACAGAGGGAGGCAGGUCGAAACGUGGAAUCAUGAAAAAUUGAUGCCCCUGUCGUCUGGUGGCGCCACCUGGUCUGCCCUCUCUCUCGCACGGCAUCUGGCCGUCUCUUUCGUGCA .......((((((((((((..((....((((.((((........))))(((((........)).))).)))).))..))).))))))))).(((((((.....)))(.......))))). ( -45.90) >consensus AAACAACAGAGGGAGGCAGGACGAAACGUGGAAUCAUGAAAAAUUGAUGCCCCUGUCGUCUGGUGGCGCCACCUGGUCUGCCCUCUCUCUCGCACGGCAUCUGGCCGUCUCUUUCGUGCA .......((((((((((((((((....((((.((((........))))(((((........)).))).)))).))))))).))))))))).(((((((.....)))(.......))))). (-42.94 = -43.50 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:18 2006