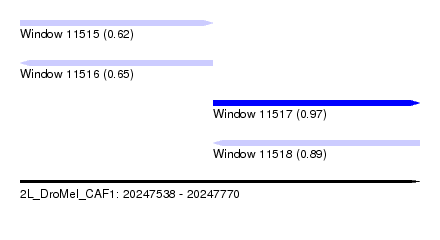
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,247,538 – 20,247,770 |
| Length | 232 |
| Max. P | 0.969115 |

| Location | 20,247,538 – 20,247,650 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -23.43 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247538 112 + 22407834 AAAAAGAGAUGAAAAUAUGAUGUGUCCCGGUCCAUAAACCAUUUGAGAAGAAUCAACAAAAAGCAAGC-----CAAGCUUCCGUUCCGAGGACAAUCAUAUUCUCCCACCGGUU---CGG .....((((.....(((((((.((((((((..(.........((((......))))....((((....-----...))))..)..))).))))))))))))))))...(((...---))) ( -26.10) >DroSec_CAF1 6072 112 + 1 AAAAAGAGAUGAAAAUAUGAUGUGUCCCGGUCCAUAAACCAUUUGAGAAGAAUCAACAAAAAGCAAGCCAAGCCAAGCUUCCGUCCCGAGGACAUUCAUAUUCGCCCAC--------CGG ..........(..(((((((.(((((((((..(.........((((......))))......(.((((........)))).))..))).)))))))))))))..)....--------... ( -25.10) >DroSim_CAF1 6503 120 + 1 AAAAAGAGAUGAAAAUAUGAUGUGUCCCGGUCCAUAAACCAUUUGAGAAGAAUCAACAAAAAGCAAGCCAAGCCAAGCUUCCGUUCCGAGGACAUUCAUAUUCUCCCACCGGUGCACCGG .....((((.....((((((.(((((((((..(.........((((......))))......(.((((........)))).))..))).))))))))))))))))...((((....)))) ( -32.10) >consensus AAAAAGAGAUGAAAAUAUGAUGUGUCCCGGUCCAUAAACCAUUUGAGAAGAAUCAACAAAAAGCAAGCCAAGCCAAGCUUCCGUUCCGAGGACAUUCAUAUUCUCCCACCGGU____CGG ..........((.(((((((.(((((((((..(.........((((......))))......(.((((........)))).))..))).))))))))))))).))............... (-23.43 = -24.10 + 0.67)

| Location | 20,247,538 – 20,247,650 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -29.63 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247538 112 - 22407834 CCG---AACCGGUGGGAGAAUAUGAUUGUCCUCGGAACGGAAGCUUG-----GCUUGCUUUUUGUUGAUUCUUCUCAAAUGGUUUAUGGACCGGGACACAUCAUAUUUUCAUCUCUUUUU (((---......)))((((((((((((((((.(((..(((((((...-----....))))))..((((......)))).........)..)))))))).))))))))))).......... ( -31.80) >DroSec_CAF1 6072 112 - 1 CCG--------GUGGGCGAAUAUGAAUGUCCUCGGGACGGAAGCUUGGCUUGGCUUGCUUUUUGUUGAUUCUUCUCAAAUGGUUUAUGGACCGGGACACAUCAUAUUUUCAUCUCUUUUU ..(--------((((..((((((((.(((((.(((..((.(((((......)))))(((.((((..((....)).)))).)))...))..))))))))..)))))))))))))....... ( -33.80) >DroSim_CAF1 6503 120 - 1 CCGGUGCACCGGUGGGAGAAUAUGAAUGUCCUCGGAACGGAAGCUUGGCUUGGCUUGCUUUUUGUUGAUUCUUCUCAAAUGGUUUAUGGACCGGGACACAUCAUAUUUUCAUCUCUUUUU ((((....))))...((((((((((.(((((.(((..((.(((((......)))))(((.((((..((....)).)))).)))...))..))))))))..)))))))))).......... ( -38.10) >consensus CCG____ACCGGUGGGAGAAUAUGAAUGUCCUCGGAACGGAAGCUUGGCUUGGCUUGCUUUUUGUUGAUUCUUCUCAAAUGGUUUAUGGACCGGGACACAUCAUAUUUUCAUCUCUUUUU ...............((((((((((.(((((.(((..((.(((((......)))))(((.((((..((....)).)))).)))...))..))))))))..)))))))))).......... (-29.63 = -30.30 + 0.67)


| Location | 20,247,650 – 20,247,770 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247650 120 + 22407834 UUCGGGGGUCCAUAAAACUCCUGGCGGGAGUGCGACCAUUUCUGGUUAAGGCUCCCACAUAUGAGCCAUAAAAGAGUAACAACAAAUACCGAAAUUGUUAUUAAAUAACAACAUAACAAU (((((.((((......(((((.....)))))..))))..((((..(((.(((((........))))).))).))))............)))))((((((((...........)))))))) ( -32.30) >DroSec_CAF1 6184 117 + 1 UUCGGGGGUCCAUAAAACUCCUGGCGGGAGUGCGACCAUUUCUGGUUAGGGCCCCCACAUAUGAGCCAUAAAAGAGUAACAACAAAUACCGAAAUUGUUAUUAAAUAA---CAUAACAAU (((((((((((.....(((((.....)))))..(((((....))))).)))))))).................(.(((........)))))))((((((((.......---.)))))))) ( -35.10) >DroSim_CAF1 6623 120 + 1 UUCGGGGGUCCAUAAAACUCCUGGCGGGAGUGCGACCAUUUCUGGUUAGGGCUCCCACAUAUGAGCCAUAAAAGAGUAACAACAAAUACCGAAAUUGUUAUUAAAUAACAACAUAACAAU (((((...........((((.((((((((((.((((((....))))).).)))))).(....).)))).....))))...........)))))((((((((...........)))))))) ( -32.95) >consensus UUCGGGGGUCCAUAAAACUCCUGGCGGGAGUGCGACCAUUUCUGGUUAGGGCUCCCACAUAUGAGCCAUAAAAGAGUAACAACAAAUACCGAAAUUGUUAUUAAAUAACAACAUAACAAU ...((((((((.....(((((.....)))))..(((((....))))).))))))))...((((...........((((((((............)))))))).........))))..... (-30.64 = -30.75 + 0.11)

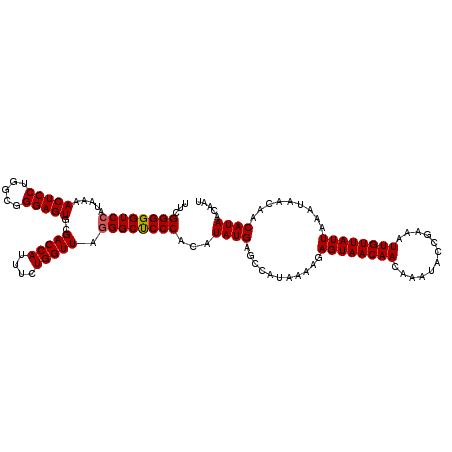
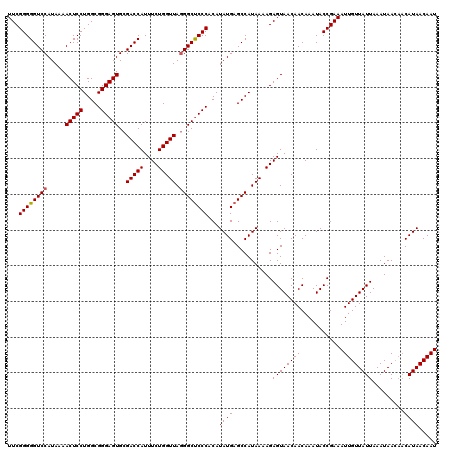
| Location | 20,247,650 – 20,247,770 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -31.92 |
| Energy contribution | -31.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20247650 120 - 22407834 AUUGUUAUGUUGUUAUUUAAUAACAAUUUCGGUAUUUGUUGUUACUCUUUUAUGGCUCAUAUGUGGGAGCCUUAACCAGAAAUGGUCGCACUCCCGCCAGGAGUUUUAUGGACCCCCGAA .......(((((((....)))))))..(((((.....((....))(((..((..((((....(((((((.....((((....))))....)))))))...))))..)).)))...))))) ( -32.00) >DroSec_CAF1 6184 117 - 1 AUUGUUAUG---UUAUUUAAUAACAAUUUCGGUAUUUGUUGUUACUCUUUUAUGGCUCAUAUGUGGGGGCCCUAACCAGAAAUGGUCGCACUCCCGCCAGGAGUUUUAUGGACCCCCGAA ...((((((---.....(((((((((.........)))))))))......)))))).......((((((.((..((((....))))...(((((.....))))).....)).)))))).. ( -33.80) >DroSim_CAF1 6623 120 - 1 AUUGUUAUGUUGUUAUUUAAUAACAAUUUCGGUAUUUGUUGUUACUCUUUUAUGGCUCAUAUGUGGGAGCCCUAACCAGAAAUGGUCGCACUCCCGCCAGGAGUUUUAUGGACCCCCGAA .......(((((((....)))))))..(((((.....((....))(((..((..((((....(((((((.....((((....))))....)))))))...))))..)).)))...))))) ( -32.00) >consensus AUUGUUAUGUUGUUAUUUAAUAACAAUUUCGGUAUUUGUUGUUACUCUUUUAUGGCUCAUAUGUGGGAGCCCUAACCAGAAAUGGUCGCACUCCCGCCAGGAGUUUUAUGGACCCCCGAA ((((((((...........))))))))(((((.....((....))(((..((..((((....(((((((.....((((....))))....)))))))...))))..)).)))...))))) (-31.92 = -31.70 + -0.22)

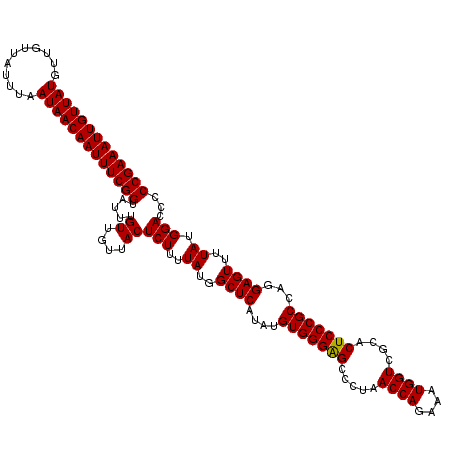
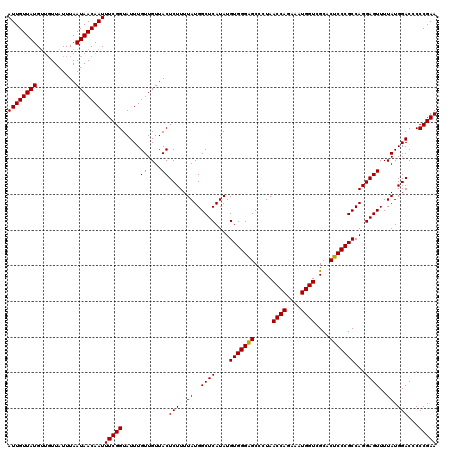
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:13 2006