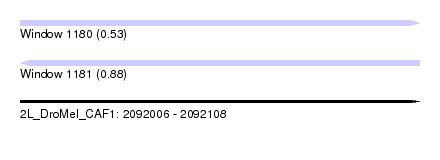
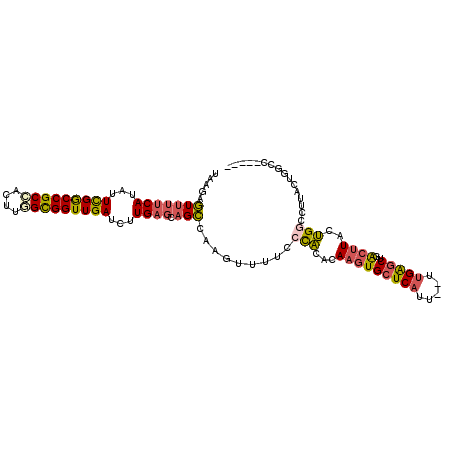
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,092,006 – 2,092,108 |
| Length | 102 |
| Max. P | 0.879563 |

| Location | 2,092,006 – 2,092,108 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -19.02 |
| Energy contribution | -17.69 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2092006 102 + 22407834 -----GUCCAGUAAGGCCAGUAAGUCAGCUCAA--AAUGAGCACUUGUGUGGGAAAACUUGGCUACUCAAGAUCAACCGCCAAGUGGCGGCCGAAUAUGAAAACUCUUA -----...((....((((.........((((..--...))))(((((.((((.....(((((....))))).....)))))))))...)))).....)).......... ( -29.80) >DroPse_CAF1 146712 109 + 1 AUGAGGGCCUGGCCGGCCCGUAGUCCUGCCCGAAUAAUGAGCACUUAUUGAGGAAAUUUAUCCGAAACAAGAUCAACCACAGUUUUGCGGUCAAAUAUGAAAAGUCCUG .((((((((.....)))))((((..(((......((((((....)))))).(((......)))................)))..))))..)))................ ( -22.80) >DroSec_CAF1 109041 102 + 1 -----GGCCAGUAAGGCCAGUAAGUCAGCUCAA--AAUGAGCACUUGUGUGGGAAAACUUGGCUGCUCAAGAUCAACCGCCAAGUGGCGGCCGAAUAUGAAAACUCCUA -----((((.....))))..(((((..((((..--...)))))))))..(((((...(((((....))))).((..(((((....)))))..))..........))))) ( -32.10) >DroSim_CAF1 108913 102 + 1 -----GGCCAGUAAGGCCAGUAAGUCAGCUCAA--AAUGAGCACUUGUGUGGGAAAACUUGGCUGCUCAAGAUCAACCGCCAAGUGGCGGCCGAAUAUGAAAACUCUUA -----((((.....))))(((...(((((((..--...)))).((((.(..(..........)..).)))).((..(((((....)))))..))...)))..))).... ( -31.70) >DroEre_CAF1 111752 97 + 1 -----GGC-----AGGUCAGUAAGUCAGCUCAA--AAUGAGCACUUGUGUGGGAAAACUUGGCUGCUCAAGAUCAACCGCAAAGUGGCGGCCGAAUAUGAAAACUCUUA -----(((-----..(((.........((((..--...))))((((.(((((.....(((((....))))).....)))))))))))).)))................. ( -26.60) >DroYak_CAF1 113638 97 + 1 -----GGC-----AGGUCACUAAGUCAGUUCAA--AAUGAGCACUUGUGUGCAAGAACUUGGCUACUCAAGAUCAACCACAAAGUGGCGGCCGAAUAUGAAAACUCUUG -----(((-----..((((((((((..((((..--...)))))))).((((...((.(((((....))))).))...)))).)))))).)))................. ( -25.30) >consensus _____GGCCAGUAAGGCCAGUAAGUCAGCUCAA__AAUGAGCACUUGUGUGGGAAAACUUGGCUACUCAAGAUCAACCGCAAAGUGGCGGCCGAAUAUGAAAACUCUUA ..............((((..(((((..(((((.....))))))))))..............((((((...............))))))))))................. (-19.02 = -17.69 + -1.33)

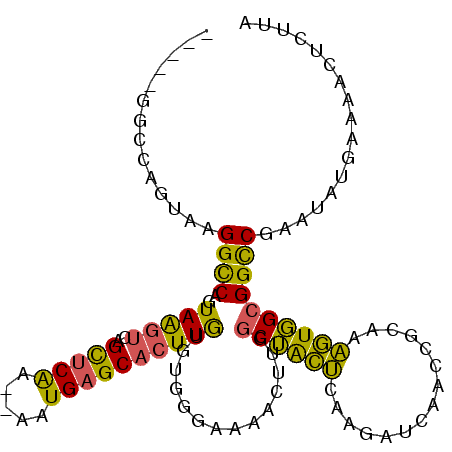
| Location | 2,092,006 – 2,092,108 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2092006 102 - 22407834 UAAGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGUAGCCAAGUUUUCCCACACAAGUGCUCAUU--UUGAGCUGACUUACUGGCCUUACUGGAC----- ........((((...(((((((((....)))))))))...))))...(((.((....(((...((((((((...--..))))..))))..)))....)))))..----- ( -31.40) >DroPse_CAF1 146712 109 - 1 CAGGACUUUUCAUAUUUGACCGCAAAACUGUGGUUGAUCUUGUUUCGGAUAAAUUUCCUCAAUAAGUGCUCAUUAUUCGGGCAGGACUACGGGCCGGCCAGGCCCUCAU .((..(.........(..((((((....))))))..).((((((..(((......)))..))))))(((((.......))))))..))..(((((.....))))).... ( -34.10) >DroSec_CAF1 109041 102 - 1 UAGGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAUU--UUGAGCUGACUUACUGGCCUUACUGGCC----- ...((((.((((...(((((((((....)))))))))...))))((((((((......(((....))).....)--))).))))))))...((((.....))))----- ( -35.80) >DroSim_CAF1 108913 102 - 1 UAAGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAUU--UUGAGCUGACUUACUGGCCUUACUGGCC----- ...((((.((((...(((((((((....)))))))))...))))((((((((......(((....))).....)--))).))))))))...((((.....))))----- ( -35.70) >DroEre_CAF1 111752 97 - 1 UAAGAGUUUUCAUAUUCGGCCGCCACUUUGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAUU--UUGAGCUGACUUACUGACCU-----GCC----- .........(((...((((((((......))))))))...)))((((...(((..........((((((((...--..))))..)))))))...))-----)).----- ( -25.70) >DroYak_CAF1 113638 97 - 1 CAAGAGUUUUCAUAUUCGGCCGCCACUUUGUGGUUGAUCUUGAGUAGCCAAGUUCUUGCACACAAGUGCUCAUU--UUGAACUGACUUAGUGACCU-----GCC----- ...(((((((((...((((((((......))))))))...))))......(((((..((((....)))).....--..))))))))))........-----...----- ( -23.60) >consensus UAAGAGUUUUCAUAUUCGGCCGCCACUUGGCGGUUGAUCUUGAGCAGCCAAGUUUUCCCACACAAGUGCUCAUU__UUGAGCUGACUUACUGGCCUUACUGGCC_____ .....(((((((...(((((((((....)))))))))...)))).))).........(((...(((((((((.....)))))..))))..)))................ (-21.63 = -21.83 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:49 2006