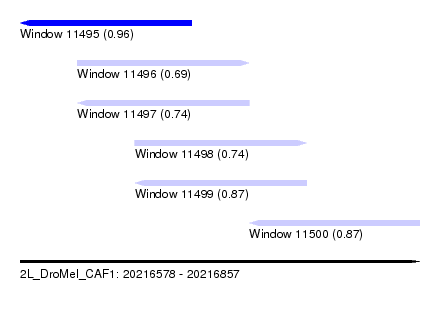
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,216,578 – 20,216,857 |
| Length | 279 |
| Max. P | 0.964025 |

| Location | 20,216,578 – 20,216,698 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216578 120 - 22407834 AUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUACUAUACAAAUAAACAGAAAUUGCGGCUAUAAAAAUGCCAA .........((((.(............).)))).(((....).......................(((((((........)))))))..............))(((.........))).. ( -18.70) >DroEre_CAF1 13316 120 - 1 AUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUACUAUACAAAUAAACAGAAAUUGCGGCUAUAAAAAUGCCAA .........((((.(............).)))).(((....).......................(((((((........)))))))..............))(((.........))).. ( -18.70) >DroYak_CAF1 10839 119 - 1 AUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUACUAUACAAAUAAACAGAAAUUGCGGCU-UAAAAAUGCCAA .........((((.(............).)))).(((....).......................(((((((........)))))))..............))(((.-.......))).. ( -19.50) >consensus AUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUACUAUACAAAUAAACAGAAAUUGCGGCUAUAAAAAUGCCAA .........((((.(............).)))).(((....).......................(((((((........)))))))..............))(((.........))).. (-18.70 = -18.70 + -0.00)

| Location | 20,216,618 – 20,216,738 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216618 120 + 22407834 UAGUAUAUCUAUACAUGUUUGUUUUUUAGUUUUCUGUUGUUUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAG ..((((....))))....................(((..((((((.(((((((((((......))))..((((((....))))))............)).))))).))))))..)))... ( -29.30) >DroEre_CAF1 13356 120 + 1 UAGUAUAUCUAUACAUGUUUGUUUUUUAGUUUUCUGUUGUUUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAG ..((((....))))....................(((..((((((.(((((((((((......))))..((((((....))))))............)).))))).))))))..)))... ( -29.30) >DroYak_CAF1 10878 120 + 1 UAGUAUAUCUAUACAUGUUUGUUUUUUAGUUUUCUGUUGUUUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAG ..((((....))))....................(((..((((((.(((((((((((......))))..((((((....))))))............)).))))).))))))..)))... ( -29.30) >consensus UAGUAUAUCUAUACAUGUUUGUUUUUUAGUUUUCUGUUGUUUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAG ..((((....))))....................(((..((((((.(((((((((((......))))..((((((....))))))............)).))))).))))))..)))... (-29.30 = -29.30 + 0.00)


| Location | 20,216,618 – 20,216,738 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216618 120 - 22407834 CUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUA .((((..(((((...(((((...............((((((....)))))).....(......).....)))))..)))))...))))..................((((....)))).. ( -18.00) >DroEre_CAF1 13356 120 - 1 CUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUA .((((..(((((...(((((...............((((((....)))))).....(......).....)))))..)))))...))))..................((((....)))).. ( -18.00) >DroYak_CAF1 10878 120 - 1 CUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUA .((((..(((((...(((((...............((((((....)))))).....(......).....)))))..)))))...))))..................((((....)))).. ( -18.00) >consensus CUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAAACAACAGAAAACUAAAAAACAAACAUGUAUAGAUAUACUA .((((..(((((...(((((...............((((((....)))))).....(......).....)))))..)))))...))))..................((((....)))).. (-18.00 = -18.00 + 0.00)


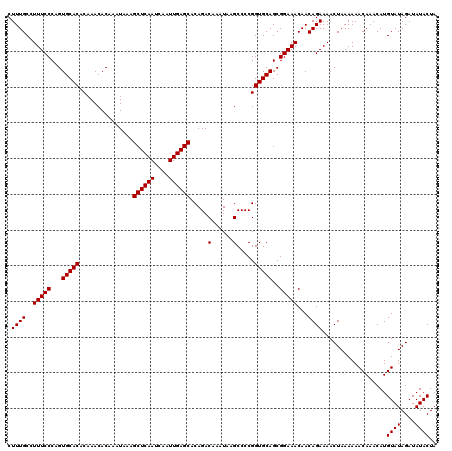
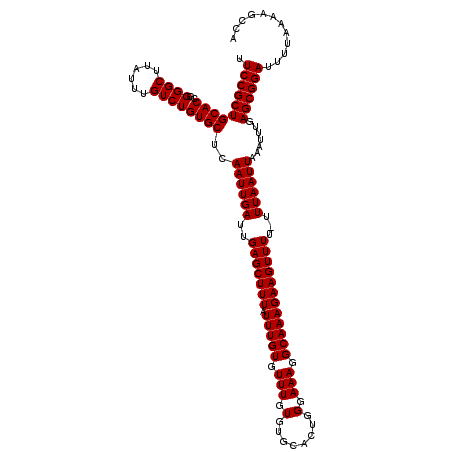
| Location | 20,216,658 – 20,216,778 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216658 120 + 22407834 UUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAGAAGUUUUUUUUAAUUAAAUUUGAGCGGAUUUUAAAAGUCA .((((((((((((((((......))))..((((((....))))))............)).))))..((((((((((......))))))))))..........))))))............ ( -31.40) >DroEre_CAF1 13396 119 + 1 UUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAGAAGUUUU-UUUAAUUAAAUUUGAGCGGAUUUUAAAAGCCA .((((((((((..((((......))))))))..((((((..(((((((.(((((.(((.(........).))).)))))))))))).-.)))))).......))))))............ ( -32.30) >DroYak_CAF1 10918 119 + 1 UUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAGAAGUUUU-UUUAAUUAAAUUUGAGCGGAUUUUAAAAGCCA .((((((((((..((((......))))))))..((((((..(((((((.(((((.(((.(........).))).)))))))))))).-.)))))).......))))))............ ( -32.30) >consensus UUCCGCUGCACCGGGGCUUAUUUGUCUGUGCUCAAUUGAUUGAGCUUUAUUUGUGUUUGUGUGCACUGGGAAAGGCAAAGAAGUUUU_UUUAAUUAAAUUUGAGCGGAUUUUAAAAGCCA .((((((((((..((((......))))))))..((((((..(((((((.(((((.(((.(........).))).))))))))))))...)))))).......))))))............ (-30.80 = -30.80 + 0.00)

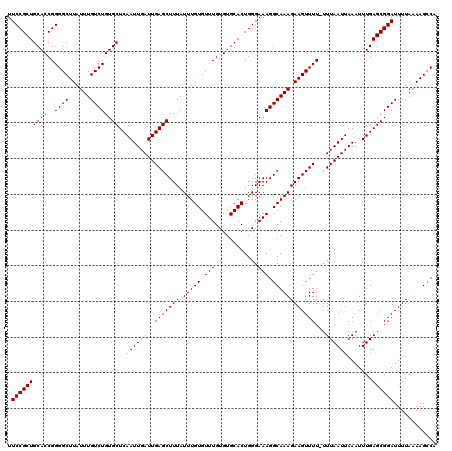
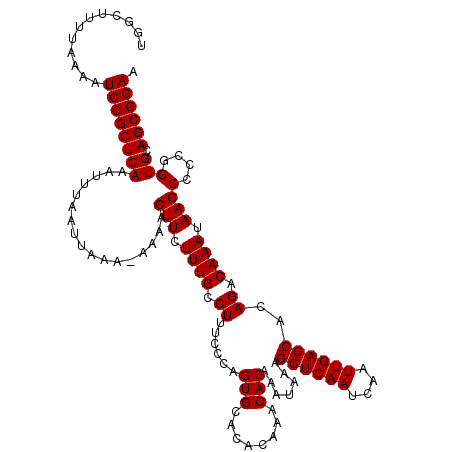
| Location | 20,216,658 – 20,216,778 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216658 120 - 22407834 UGACUUUUAAAAUCCGCUCAAAUUUAAUUAAAAAAAACUUCUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAA ............((((((((.................(((.((((.((......(((........))).......((((((....))))))..)).)))).)))(....))).)))))). ( -21.20) >DroEre_CAF1 13396 119 - 1 UGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA-AAAACUUCUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAA ............((((((((............-....(((.((((.((......(((........))).......((((((....))))))..)).)))).)))(....))).)))))). ( -21.20) >DroYak_CAF1 10918 119 - 1 UGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA-AAAACUUCUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAA ............((((((((............-....(((.((((.((......(((........))).......((((((....))))))..)).)))).)))(....))).)))))). ( -21.20) >consensus UGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA_AAAACUUCUUUGCCUUUCCCAGUGCACACAAACACAAAUAAAGCUCAAUCAAUUGAGCACAGACAAAUAAGCCCCGGUGCAGCGGAA ............((((((((.................(((.((((.((......(((........))).......((((((....))))))..)).)))).)))(....))).)))))). (-21.20 = -21.20 + -0.00)

| Location | 20,216,738 – 20,216,857 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20216738 119 - 22407834 UAAUUAAACCAAAACGCUACAAUUGCGGAUAUAGGAAAGGCAAACAGUCCUUUCCC-CCAAAGAGCAUCGCACAAUCAUCUGACUUUUAAAAUCCGCUCAAAUUUAAUUAAAAAAAACUU ((((((((................((((((...((((((((.....).))))))).-...(((((((.............)).)))))...)))))).....)))))))).......... ( -19.82) >DroEre_CAF1 13476 118 - 1 UAAUUAAACCAAAACGUUGCAAUUGCGGAUAUAGAAAAGGCAAACCGUCCUUUUCC-CCAAAGAGCAUCACACAAUCAUCUGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA-AAAACUU ((((((((.(((....))).....((((((...((((((((.....).))))))).-...((((((................))))))...)))))).....))))))))..-....... ( -21.59) >DroYak_CAF1 10998 119 - 1 UAAUUAAACCAAAAAGUUGCAACUGCGGAUAUAGAAAAGGCAAACAGUCAUUUUCCCCCAAAGAGCAUCACACAAUCAUCUGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA-AAAACUU ((((((((......(((....)))((((((...((((((((.....))).))))).....((((((................))))))...)))))).....))))))))..-....... ( -19.99) >consensus UAAUUAAACCAAAACGUUGCAAUUGCGGAUAUAGAAAAGGCAAACAGUCCUUUUCC_CCAAAGAGCAUCACACAAUCAUCUGGCUUUUAAAAUCCGCUCAAAUUUAAUUAAA_AAAACUU ((((((((................((((((...((((((((.....).))))))).....((((((................))))))...)))))).....)))))))).......... (-17.17 = -17.39 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:57 2006