
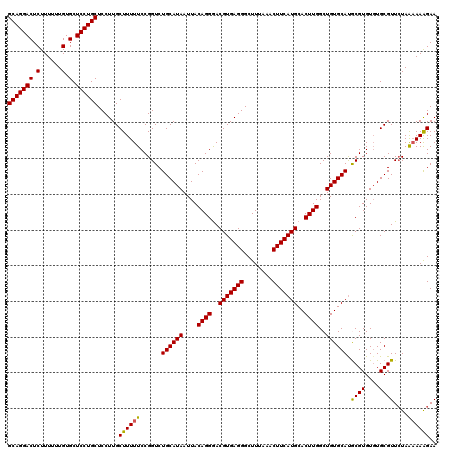
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,213,667 – 20,213,819 |
| Length | 152 |
| Max. P | 0.987459 |

| Location | 20,213,667 – 20,213,787 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
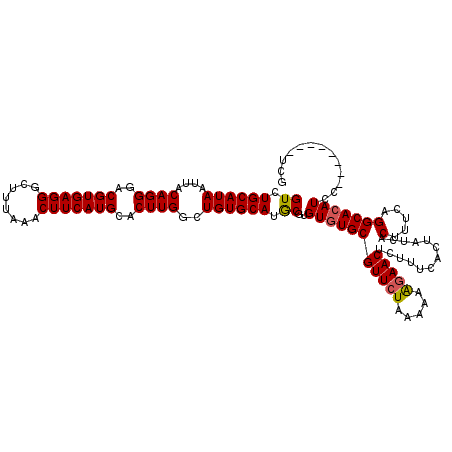
>2L_DroMel_CAF1 20213667 120 + 22407834 GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCACGCGUGUGUGCGUUCUAAAAAAGAA (((((((.(......).).))))))......((((((..((...(((((....((((..(((((((........)))))))..))))..)))))(((((....)))))))..)))))).. ( -35.90) >DroSec_CAF1 54880 120 + 1 GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGAACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAGGAA (((((((.(......).).))))))(((((((............)).........(((((((((((........))))((((((......))))))........)))))))...))))). ( -35.60) >DroSim_CAF1 32052 120 + 1 GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAAGAA (((((((.(......).).))))))......((((((..((..((((((....((((..(((((((........)))))))..))))((.......))...))))))..)).)))))).. ( -35.50) >DroEre_CAF1 10332 120 + 1 GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUACGUGUUUGCGUUCUAAAAAAGAA (((((((.(......).).))))))......((((((..((..((((((....((((..(((((((........)))))))..))))..)))))).((((....))))))..)))))).. ( -34.40) >DroYak_CAF1 8070 119 + 1 GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUACGUGUGUGCGUUAUAG-AAGGAA (((((((.(......).).))))))(((((.............((((((....((((..(((((((........)))))))..))))..)))))).((((....)))).....-))))). ( -36.90) >consensus GCAGGACUCUUUUUUGUGCUCCUGCUCCUUGCUUUUUCCGGUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAAGAA (((((((.(......).).))))))......((((((......((((((....((((..(((((((........)))))))..))))..)))))).((((....))))....)))))).. (-33.84 = -33.40 + -0.44)


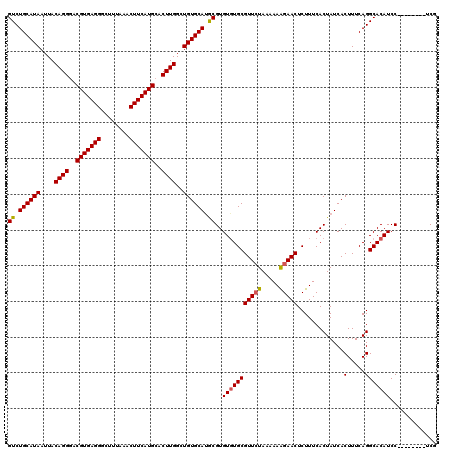
| Location | 20,213,707 – 20,213,819 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20213707 112 + 22407834 GUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCACGCGUGUGUGCGUUCUAAAAAAGAACUCUUUCACUAUCACUUUCAGGCACAUCC--------UCG ((.((((((....((((..(((((((........)))))))..))))..)))))).))(..((((((((((.....))))).............(.....))))))..)--------... ( -33.10) >DroSec_CAF1 54920 112 + 1 GUCUGCAUAAUUACAGGAACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAGGAACUCUUUCACUAUCACUUUCAGGCACAUCC--------UCG ((.((((((....((((..(((((((........)))))))..))))..)))))).))(..((((((((((.....))))).............(.....))))))..)--------... ( -31.10) >DroSim_CAF1 32092 112 + 1 GUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAAGAACUCUUUCACUAUCACUUUCAGGCACAUCC--------UCG ((.((((((....((((..(((((((........)))))))..))))..)))))).))(..((((((((((.....))))).............(.....))))))..)--------... ( -33.10) >DroEre_CAF1 10372 120 + 1 GUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUACGUGUUUGCGUUCUAAAAAAGAACUCUUUCACUAUCACUUUCAGGCACAUCCUCGCAUCCUCG ..(((........)))(((.((((((((((.(((....((((((......))))))..(((...(.(((((.....))))).)...)))......))).))))....)))))).)))... ( -32.40) >DroYak_CAF1 8110 111 + 1 GUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUACGUGUGUGCGUUAUAG-AAGGAACUCUUUCACUAUCACUUUCAGGCACAUCC--------UCG ((.((((((....((((..(((((((........)))))))..))))..)))))).))(..(((((......(-((((....))))).......(.....))))))..)--------... ( -33.20) >consensus GUCUGCAUAAUUACAGGGACGUGAGGGCUUUAAACUUCAUGCACUUGGCUGUGCAUGCGUGUGUGCGUUCUAAAAAAGAACUCUUUCACUAUCACUUUCAGGCACAUCC________UCG ((.((((((....((((..(((((((........)))))))..))))..)))))).))..(((((((((((.....))))).............(.....)))))))............. (-30.10 = -30.02 + -0.08)

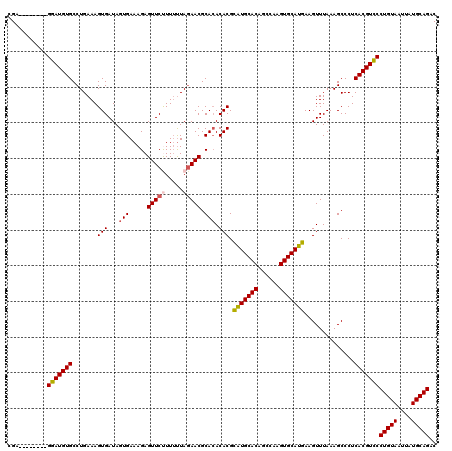
| Location | 20,213,707 – 20,213,819 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -28.62 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20213707 112 - 22407834 CGA--------GGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCUUUUUUAGAACGCACACACGCGUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUCCCUGUAAUUAUGCAGAC ...--------(((((((.......(((...(((.....(((((.....))))).))).))).(((((((......)))))))..............)))))))(((((....))))).. ( -32.30) >DroSec_CAF1 54920 112 - 1 CGA--------GGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCCUUUUUAGAACGCACACACGCAUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUUCCUGUAAUUAUGCAGAC .((--------((.((((((((((((.((...(....)...)).)))))))...)))))....(((((((......)))))))...........))))......(((((....))))).. ( -31.40) >DroSim_CAF1 32092 112 - 1 CGA--------GGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCUUUUUUAGAACGCACACACGCAUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUCCCUGUAAUUAUGCAGAC ...--------(((((((.......(((...(((.....(((((.....))))).))).))).(((((((......)))))))..............)))))))(((((....))))).. ( -32.70) >DroEre_CAF1 10372 120 - 1 CGAGGAUGCGAGGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCUUUUUUAGAACGCAAACACGUAUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUCCCUGUAAUUAUGCAGAC ...(((((.((((..(((.(((((((.((...(....)...)).)))))))..)))((((...(((((((......)))))))..)))).....)))).)))))(((((....))))).. ( -35.90) >DroYak_CAF1 8110 111 - 1 CGA--------GGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCCUU-CUAUAACGCACACACGUAUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUCCCUGUAAUUAUGCAGAC ...--------(((((((.......(((...((...((((....))-))...)).))).....(((((((......)))))))..............)))))))(((((....))))).. ( -27.40) >consensus CGA________GGAUGUGCCUGAAAGUGAUAGUGAAAGAGUUCUUUUUUAGAACGCACACACGCAUGCACAGCCAAGUGCAUGAAGUUUAAAGCCCUCACGUCCCUGUAAUUAUGCAGAC ...........(((((((.......(((...(((.....(((((.....))))).))).))).(((((((......)))))))..............)))))))(((((....))))).. (-28.62 = -28.86 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:51 2006