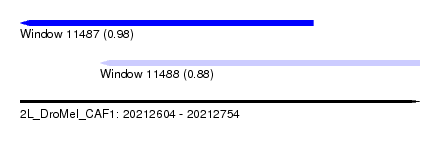
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,212,604 – 20,212,754 |
| Length | 150 |
| Max. P | 0.979547 |

| Location | 20,212,604 – 20,212,714 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -28.46 |
| Energy contribution | -29.14 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20212604 110 - 22407834 AACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAUGUGUUCCUAAUUUCGAUAAAGGCAUUACCCAGAUAUGGA---------- ...........((((.........(..(((....)))..)...((((((((((..(((.(((.((.(((....)))))))).)))..))))))))))..)))).......---------- ( -34.50) >DroSec_CAF1 53837 110 - 1 AACGCUUGCAACUGGAACAACGAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUAAUUUCGAUAAAGGCAUUGCCCAGAUAUGGA---------- ...........((((.........(..(((....)))..)...((((((((((..(((.(((.((.(((....)))))))).)))..))))))))))..)))).......---------- ( -34.50) >DroSim_CAF1 31009 110 - 1 AACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGGAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUAAUUUCGAUAAAGGCAUUGCCCAGAUAUGGA---------- .((((((((.(((..........)))....))))))))((...((((((((((..(((.(((.((.(((....)))))))).)))..))))))))))..)).........---------- ( -31.90) >DroEre_CAF1 9379 120 - 1 CACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCGAGUGUUCCUAAUUUCGGUAAAGGCAUUACCGAGAUAUGGUUUCUGUUCCA .............((((((.....(..(((....)))..)...((((((((((((((((((((....)))))...)))).......)))))))))))...(((((...))))))))))). ( -39.91) >DroYak_CAF1 7031 119 - 1 UACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUACAUAAAUGCCUUUGCAGAACUUGUGGC-GCAAGUGUUCCUAAUUUCGAUAAAGACAUUACCGAGAUAUGGUUUCUGUUCUA .((((((((...............((((((....))))))......(((...((((...)))))))-)))))))).((..((((((.(((.....))).))))))..))........... ( -32.40) >consensus AACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUAAUUUCGAUAAAGGCAUUACCCAGAUAUGGA__________ ...........((((.........((((((....))))))...((((((((((..(((.(((.((.(((....)))))))).)))..))))))))))..))))................. (-28.46 = -29.14 + 0.68)


| Location | 20,212,634 – 20,212,754 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -33.05 |
| Energy contribution | -32.81 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20212634 120 - 22407834 CCUCGUUGCGACUCGACCAGAGUUAAUGUGCCGCGAUUUUAACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAUGUGUUCCUA ....((((((((((.....)))))........(((.......)))..))))).((((((.....(..(((....)))..)....((((..(..(((...)))..)..)))).)))))).. ( -39.40) >DroSec_CAF1 53867 120 - 1 CCUCUUGGCGACUCGACCAGAGUUAAUGUGCCGCGAUUUUAACGCUUGCAACUGGAACAACGAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUA ..((.(((((((((.....))))).....)))).))....(((((((((...............(..(((....)))..).....((((...((((...))))))))))))))))).... ( -37.70) >DroSim_CAF1 31039 120 - 1 CCUCGUUGCGACUCGACCAGAGUUAAUGUGCCGCGAUUUUAACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGGAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUA ....((((((((((.....)))))........(((.......)))..))))).((((((......(((.((((((((..((........))..)...)))))))))).....)))))).. ( -35.90) >DroEre_CAF1 9419 120 - 1 CCUCGUUGGGACUCUGGCAGAGUUAAUGUGCCGCGAUUUUCACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCGAGUGUUCCUA .((((((..(((((.....))))).....(((((((........(((((((..((.........(..(((....)))..).......)).)))))))..)))))))))))))........ ( -39.69) >DroYak_CAF1 7071 119 - 1 CCUCGUUGGAACUCAGCCAGAGUUAAUGCGCCGCGAUUUUUACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUACAUAAAUGCCUUUGCAGAACUUGUGGC-GCAAGUGUUCCUA .......(((((.((((....)))..((((((((((.(((......(((((..((.........((((((....)))))).......)).)))))))).)))))))-))).).))))).. ( -39.89) >consensus CCUCGUUGCGACUCGACCAGAGUUAAUGUGCCGCGAUUUUAACGCUUGCAACUGGAACAACAAAGUGCACGCAAGUGUGCAUAAAUGCCUUUGCAGGACUUGUGGCAGCAAGUGUUCCUA .........(((((.....)))))..((((((((((........(((((((..((.........((((((....)))))).......)).)))))))..))))))).))).......... (-33.05 = -32.81 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:47 2006