
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,141,870 – 20,142,097 |
| Length | 227 |
| Max. P | 0.995890 |

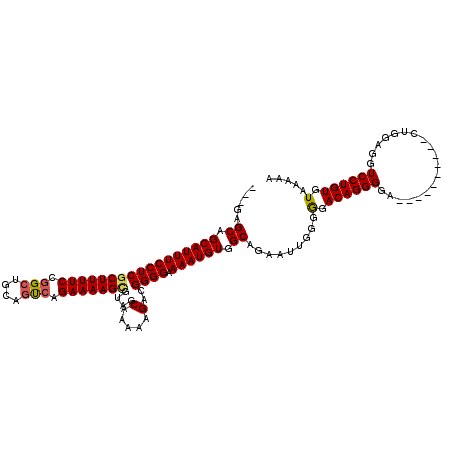
| Location | 20,141,870 – 20,141,977 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -26.87 |
| Energy contribution | -26.95 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141870 107 + 22407834 ---GAGCAGCAUUUCCUCGCUUUUCCGGCUGCAGUCAGAAAAGCGUAGCAAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGAA----------CUGGAGGUCCUGUGUAAAAA ---..((.((((((((((((((((...(((((.((.......))))))).))))).))))).)))))).)).......((((.(((....----------)))....))))......... ( -34.30) >DroSec_CAF1 3754 107 + 1 ---GAGCAGCAUUUCCUCCCUUUUCCGGCUGCAGUCAGAAAAGCGUAGCGAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGGA----------CUGGAGGUCCUGUGUAAAAA ---..((.((((((((((.((((((..(((((.((.......)))))))))))))..)))).)))))).))...........(((.((((----------(.....))))).)))..... ( -35.30) >DroSim_CAF1 5421 110 + 1 GAGGAGCAGCAUUUCCUCGCUUUUCCGGCUGCAGUCAGAAAAGCGUAGCGAAAAGACGGGGAAAAUGUGGCAGAAUGGGGGGACAGGGGA----------CUGGAGGUCCUGUGUAAAAA .....((.(((((((((((((((((..(((((.((.......))))))))))))).))))).)))))).))...........(((.((((----------(.....))))).)))..... ( -39.50) >DroEre_CAF1 5162 115 + 1 ---CAGCAGCAUUUCCUCGCUUUUCCGGCUGCAACGAGAAAAGCGAAGCAAAAAGA-GGGGAAAAUGUGGCGGAAU-GGGGGACAGGGGGUGCGGGGCUGCAGGAGGUCCUGUGUAAAAA ---.(((.((((..(((((((((((((.......)).)))))))))..........-...................-.(....).))..))))...)))(((((....)))))....... ( -38.60) >DroYak_CAF1 2930 116 + 1 ---GAGCAGCAUUUCCUCGCUUUUCCGGCUGCAGCCAGAAAAGUGAAGCAAAAUGA-GGGGAAAAUGUGGCUGAAUUAGGAGACAGGGGGUUCGGGGAUGCAUGAGGUCCUGUGUAAAAA ---.(((.(((((((((((((((((.(((....))).)))))))....(.....).-)))).)))))).)))......(....).((((.((((........)))).))))......... ( -34.60) >consensus ___GAGCAGCAUUUCCUCGCUUUUCCGGCUGCAGUCAGAAAAGCGUAGCAAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGGA__________CUGGAGGUCCUGUGUAAAAA .....((.(((((((((((((((((.(((....))).)))))))....(.....)..)))).)))))).)).........(.((((((...................)))))).)..... (-26.87 = -26.95 + 0.08)

| Location | 20,141,870 – 20,141,977 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -19.16 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141870 107 - 22407834 UUUUUACACAGGACCUCCAG----------UUCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUUGCUACGCUUUUCUGACUGCAGCCGGAAAAGCGAGGAAAUGCUGCUC--- .......(((((........----------...)))))...........((..(((((((...((......))..((((((((((........)))))))))).)))))))..))..--- ( -24.70) >DroSec_CAF1 3754 107 - 1 UUUUUACACAGGACCUCCAG----------UCCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUCGCUACGCUUUUCUGACUGCAGCCGGAAAAGGGAGGAAAUGCUGCUC--- .....(((..((((.....)----------)))..)))...........((..((((((((((...((((((((..((..........)))).)))))).))).)))))))..))..--- ( -25.10) >DroSim_CAF1 5421 110 - 1 UUUUUACACAGGACCUCCAG----------UCCCCUGUCCCCCCAUUCUGCCACAUUUUCCCCGUCUUUUCGCUACGCUUUUCUGACUGCAGCCGGAAAAGCGAGGAAAUGCUGCUCCUC .....(((..((((.....)----------)))..)))...........((..(((((((..((......))...((((((((((........)))))))))).)))))))..))..... ( -26.60) >DroEre_CAF1 5162 115 - 1 UUUUUACACAGGACCUCCUGCAGCCCCGCACCCCCUGUCCCCC-AUUCCGCCACAUUUUCCCC-UCUUUUUGCUUCGCUUUUCUCGUUGCAGCCGGAAAAGCGAGGAAAUGCUGCUG--- .........(((....)))(((((...(((.....))).....-...................-........((((((((((((.((....)).))))))))))))....)))))..--- ( -27.80) >DroYak_CAF1 2930 116 - 1 UUUUUACACAGGACCUCAUGCAUCCCCGAACCCCCUGUCUCCUAAUUCAGCCACAUUUUCCCC-UCAUUUUGCUUCACUUUUCUGGCUGCAGCCGGAAAAGCGAGGAAAUGCUGCUC--- .......(((((.....................)))))..........(((..(((((((...-..........((.((((((((((....)))))))))).)))))))))..))).--- ( -26.41) >consensus UUUUUACACAGGACCUCCAG__________UCCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUUGCUACGCUUUUCUGACUGCAGCCGGAAAAGCGAGGAAAUGCUGCUC___ .......(((((.....................)))))...........((..(((((((..((......))...((((((((((........)))))))))).)))))))..))..... (-19.16 = -20.08 + 0.92)


| Location | 20,141,907 – 20,142,017 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -25.93 |
| Energy contribution | -26.69 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141907 110 + 22407834 AAGCGUAGCAAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGAA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA ..((...((....((((((((........(((.((((...(.((((((..----------.......)))))).)...)))).))).)))))))).))...((((((....)))))))). ( -31.20) >DroSec_CAF1 3791 110 + 1 AAGCGUAGCGAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGGA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA ..(((((((((.(((((............(((.((((.....(((.((((----------(.....))))).)))...)))).)))(((......))))))))....)))))))...)). ( -36.80) >DroSim_CAF1 5461 110 + 1 AAGCGUAGCGAAAAGACGGGGAAAAUGUGGCAGAAUGGGGGGACAGGGGA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA ..(((((((((.(((((............(((.(((......(((.((((----------(.....))))).)))....))).)))(((......))))))))....)))))))...)). ( -35.70) >DroEre_CAF1 5199 118 + 1 AAGCGAAGCAAAAAGA-GGGGAAAAUGUGGCGGAAU-GGGGGACAGGGGGUGCGGGGCUGCAGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA .(((((.(((......-........)))(.((((((-(..((((((((.((((((((((......))))))))...........)).))))))))..))))))).).)))))........ ( -36.04) >DroYak_CAF1 2967 119 + 1 AAGUGAAGCAAAAUGA-GGGGAAAAUGUGGCUGAAUUAGGAGACAGGGGGUUCGGGGAUGCAUGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA .......((.(((..(-(((((..(((((.(((((((.(....)....)))))))...)))))....))).(((((.....))))).)))..))).))((((.((((....)))))))). ( -33.30) >consensus AAGCGUAGCAAAAAGACGGGGAAAAUGUGGCAGAAUUGGGGGACAGGGGA__________CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCA ..((...((....((((((((........(((.((((...(.((((((...................)))))).)...)))).))).)))))))).))...((((((....)))))))). (-25.93 = -26.69 + 0.76)


| Location | 20,141,907 – 20,142,017 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -19.28 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141907 110 - 22407834 UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UUCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUUGCUACGCUU .((.((((((.((((.......(((......)))(((.((((.....(((((........----------...))))).....)))).)))...........))))...)))))).)).. ( -23.90) >DroSec_CAF1 3791 110 - 1 UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UCCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUCGCUACGCUU .((((((((....))))))...(((......)))(((.((((...(((..((((.....)----------)))..))).....)))).)))....................))....... ( -22.70) >DroSim_CAF1 5461 110 - 1 UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UCCCCUGUCCCCCCAUUCUGCCACAUUUUCCCCGUCUUUUCGCUACGCUU .((.((((((.((((......((((......))))..........(((..((((.....)----------)))..))).....)))))))))).........((......))....)).. ( -24.10) >DroEre_CAF1 5199 118 - 1 UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCUGCAGCCCCGCACCCCCUGUCCCCC-AUUCCGCCACAUUUUCCCC-UCUUUUUGCUUCGCUU ........(((((.((.((((((((......))))............(((((......(((......)))...))))).....-...................-..)))).)).))))). ( -22.30) >DroYak_CAF1 2967 119 - 1 UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCAUGCAUCCCCGAACCCCCUGUCUCCUAAUUCAGCCACAUUUUCCCC-UCAUUUUGCUUCACUU .((((((((....))))))...(((......)))((..((((.....(((((.....................))))).....))))..))............-.......))....... ( -19.10) >consensus UGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG__________UCCCCUGUCCCCCAAUUCUGCCACAUUUUCCCCGUCUUUUUGCUACGCUU .((((((((....))))))...(((......)))(((.((((.....(((((.....................))))).....)))).)))....................))....... (-19.28 = -20.08 + 0.80)

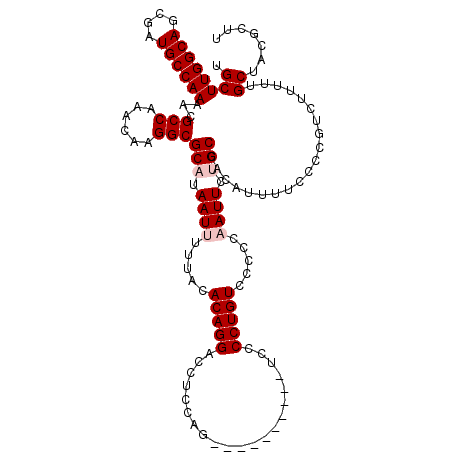
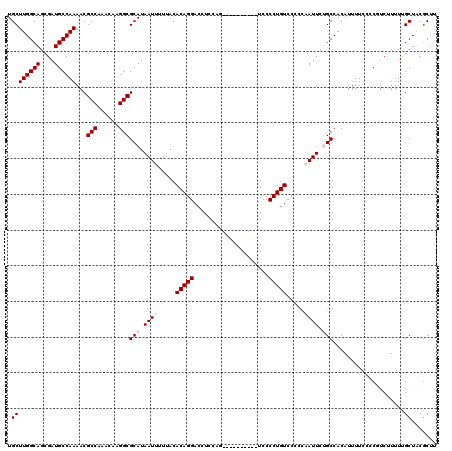
| Location | 20,141,947 – 20,142,057 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -30.98 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141947 110 + 22407834 GGACAGGGAA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAU ..((((((..----------......(((.((((((........(((((......)))))(((((((....))))))).....)))))).)))...(((((....))))))))))).... ( -33.00) >DroSec_CAF1 3831 110 + 1 GGACAGGGGA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUGU ..(((.((((----------(.....))))).)))....(((.((((((......)))...((((((....))))))))).)))((((((((.((.(((((....))))))))))))))) ( -39.60) >DroSim_CAF1 5501 110 + 1 GGACAGGGGA----------CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAU ..(((.((((----------(.....))))).)))....(((.((((((......)))...((((((....))))))))).)))..((((((.((.(((((....))))))))))))).. ( -36.10) >DroEre_CAF1 5237 120 + 1 GGACAGGGGGUGCGGGGCUGCAGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACAUACGGCAGAUGAUUAUGAGAUCACUUUUUGUAU ..(((((((((((((((((......))))))(((((........(((((......)))))(((((((....))))))).....)))))...))....((((....))))))))))))).. ( -36.90) >DroYak_CAF1 3006 120 + 1 AGACAGGGGGUUCGGGGAUGCAUGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUUUGUAU ..(((((((((((...(((.(((...(((.((((((........(((((......)))))(((((((....))))))).....)))))).)))..))))))....))..))))))))).. ( -35.10) >consensus GGACAGGGGA__________CUGGAGGUCCUGUGUAAAAAUUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAU (.((((((..................(((.((((((........(((((......)))))(((((((....))))))).....)))))).)))...(((((....))))))))))).).. (-30.98 = -31.06 + 0.08)


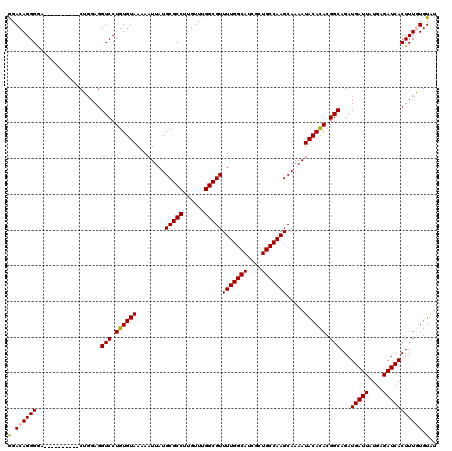
| Location | 20,141,947 – 20,142,057 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141947 110 - 22407834 AUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UUCCCUGUCC (((((((.((((((......)))).))....)))))))..(((((((((....))))))...(((......))))))..........(((((........----------...))))).. ( -28.70) >DroSec_CAF1 3831 110 - 1 ACACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UCCCCUGUCC ((((((..((((((......)))).))...))))))....(((((((((....))))))...(((......))))))........(((..((((.....)----------)))..))).. ( -31.00) >DroSim_CAF1 5501 110 - 1 AUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG----------UCCCCUGUCC (((((((.((((((......)))).))....)))))))..(((((((((....))))))...(((......))))))........(((..((((.....)----------)))..))).. ( -30.00) >DroEre_CAF1 5237 120 - 1 AUACAAAAAGUGAUCUCAUAAUCAUCUGCCGUAUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCUGCAGCCCCGCACCCCCUGUCC .........(((((......))))).(((.(((...(((.(((((((((....))))))...(((......)))))).)))...))).((((....)))).......))).......... ( -27.60) >DroYak_CAF1 3006 120 - 1 AUACAAAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCAUGCAUCCCCGAACCCCCUGUCU .........(((((......)))))...(((((((.(((.(((((((((....))))))...(((......)))))).)))...))))).))............................ ( -25.10) >consensus AUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAAUUUUUACACAGGACCUCCAG__________UCCCCUGUCC (((((((.((((((......)))).))....)))))))..(((((((((....))))))...(((......))))))..........(((((.....................))))).. (-25.20 = -25.80 + 0.60)

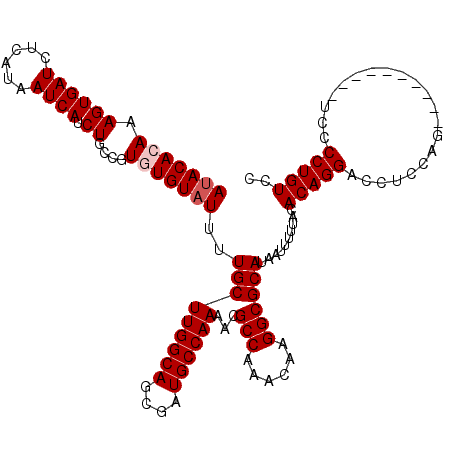

| Location | 20,141,977 – 20,142,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141977 120 + 22407834 UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAUGACUUGCUGCGAAAGCAGAUCCUUUUACCUUAAAUCCUCA ....(((((......))))).((((((....))))))((((.(((((((.((....(((((....))))))).)))))))...))))(((....)))....................... ( -34.40) >DroSec_CAF1 3861 120 + 1 UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUGUGACUUGCUGCGAAAGCAGAUCCUUUUACCUUAACUCCUCA ....(((((......))))).((((((....))))))((((..(((((((((.((.(((((....))))))))))))))))..))))(((....)))....................... ( -39.20) >DroSim_CAF1 5531 120 + 1 UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAUGACUUGCUGCGAAAGCAGAUCCUUUUACCUUAACUCCUCA ....(((((......))))).((((((....))))))((((.(((((((.((....(((((....))))))).)))))))...))))(((....)))....................... ( -34.40) >DroEre_CAF1 5277 120 + 1 UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACAUACGGCAGAUGAUUAUGAGAUCACUUUUUGUAUGACUUGCUGGAAGAGCAGAUCCUUUUACCUUAGCUCCUCA ....(((((......)))))..(((((....)))))(((((....(((((((.((.(((((....))))).)).)))))))..))))).((.((((................)))).)). ( -32.39) >DroYak_CAF1 3046 120 + 1 UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUUUGUAUGACUGGCUGGAAGAGCAGAUCCUUUUACCUUAGCUCUAUA ....(((((......)))))((..(((((((((((...............))))).))))((..(((.....)))..))......))..))(((((................)))))... ( -29.25) >consensus UUAUGCGCCUUGUUUGGCGUUUUGGCAUCGCUGCCAAGCAAAAUACACACGGCAGAUGAUUAUGAGAUCACUUUGUGUAUGACUUGCUGCGAAAGCAGAUCCUUUUACCUUAACUCCUCA ...((((((......)))...((((((....)))))))))..(((((((.((....(((((....))))))).)))))))......((((....))))...................... (-28.00 = -28.64 + 0.64)


| Location | 20,141,977 – 20,142,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20141977 120 - 22407834 UGAGGAUUUAAGGUAAAAGGAUCUGCUUUCGCAGCAAGUCAUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA ..........((((((((.((..((((.....))))..)).((((((.((((((......)))).))....))))))))))))))((((....))))....((((......))))..... ( -32.20) >DroSec_CAF1 3861 120 - 1 UGAGGAGUUAAGGUAAAAGGAUCUGCUUUCGCAGCAAGUCACACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA ......((.((((((........)))))).)).(((((..((((((..((((((......)))).))...))))))..)))))((((((....))))))..((((......))))..... ( -35.00) >DroSim_CAF1 5531 120 - 1 UGAGGAGUUAAGGUAAAAGGAUCUGCUUUCGCAGCAAGUCAUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA ......((.((((((........)))))).)).(((((..((((((..((((((......)))).))...))))))..)))))((((((....))))))..((((......))))..... ( -33.10) >DroEre_CAF1 5277 120 - 1 UGAGGAGCUAAGGUAAAAGGAUCUGCUCUUCCAGCAAGUCAUACAAAAAGUGAUCUCAUAAUCAUCUGCCGUAUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA .(((((((..((((......)))))))))))..(((((.(((((...(..((((......))))..)...)))))...)))))((((((....))))))..((((......))))..... ( -36.20) >DroYak_CAF1 3046 120 - 1 UAUAGAGCUAAGGUAAAAGGAUCUGCUCUUCCAGCCAGUCAUACAAAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA ...(((((..((((......)))))))))....((.((..(((((...((((((......)))).))....)))))..)).))((((((....))))))..((((......))))..... ( -28.80) >consensus UGAGGAGUUAAGGUAAAAGGAUCUGCUUUCGCAGCAAGUCAUACACAAAGUGAUCUCAUAAUCAUCUGCCGUGUGUAUUUUGCUUGGCAGCGAUGCCAAAACGCCAAACAAGGCGCAUAA ...(((((..((((......)))))))))....(((((..((((((..((((((......)))).))...))))))..)))))((((((....))))))..((((......))))..... (-29.28 = -29.48 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:41 2006