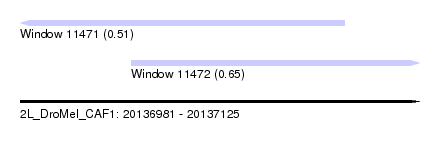
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,136,981 – 20,137,125 |
| Length | 144 |
| Max. P | 0.654582 |

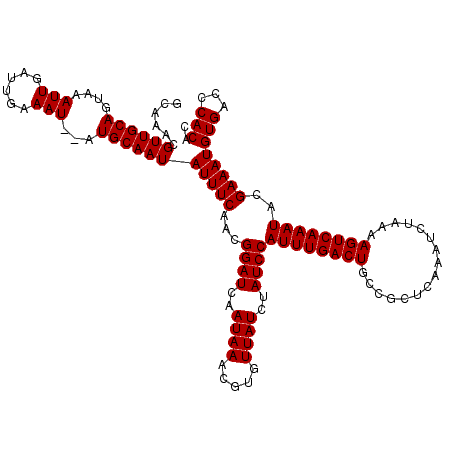
| Location | 20,136,981 – 20,137,098 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.19 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20136981 117 - 22407834 -CAAACGUUGCAGUAAAUUGAUUGAAAU--AUGCAAUAUUUCAACGGAUCAAUAAACGUGUUAUCUAUCCAUUUGACUGCCGCUCAAAUCUAAAAGUCAAAUACGAAAUGUGAACCACAC -...(((((...(((......(((((((--((...))))))))).((((..((((.....))))..)))).(((((((................))))))))))..)))))......... ( -19.69) >DroSec_CAF1 48282 118 - 1 GCAAACGUUGCAGUAAAUUGAUUGAAAU--GUGCAAUAUUUCAACGGAUCAAUAAACGUGUUAUCUAUCCAUUUGACUGCCGCUCAAAUCUAAAAGUCAAAUACGAAAUGUGACCCACAC ((..(((((.((((......)))).)))--)))).(((((((...((((..((((.....))))..))))((((((((................))))))))..)))))))......... ( -22.89) >DroYak_CAF1 6914 120 - 1 GAAAACGUUGCAGUAAAUUGAUUGAAAUAUAUGCAAUAUUUCAACGGAUCAAUAAACGUGUUAUCUAUCCAUUUGACUGCAACUCAAAUCUAAAAGUCAAAUAGGAAAUGUGACCCACAC ......((((((((.......(((((((((.....))))))))).((((..((((.....))))..)))).....))))))))............((((.((.....)).))))...... ( -25.20) >consensus GCAAACGUUGCAGUAAAUUGAUUGAAAU__AUGCAAUAUUUCAACGGAUCAAUAAACGUGUUAUCUAUCCAUUUGACUGCCGCUCAAAUCUAAAAGUCAAAUACGAAAUGUGACCCACAC ......((((((....(((......)))...))))))(((((...((((..((((.....))))..))))((((((((................))))))))..)))))(((...))).. (-20.19 = -20.19 + -0.00)

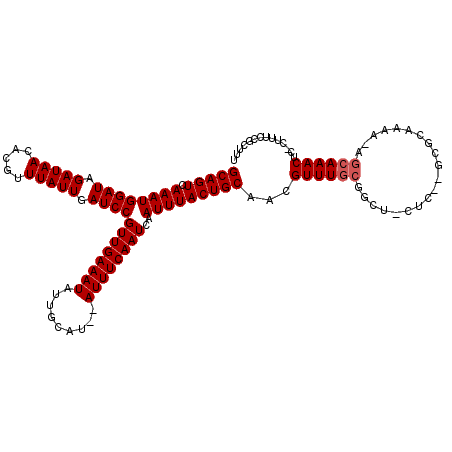
| Location | 20,137,021 – 20,137,125 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.39 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.55 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20137021 104 + 22407834 GCAGUCAAAUGGAUAGAUAACACGUUUAUUGAUCCGUUGAAAUAUUGCAU--AUUUCAAUCAAUUUACUGCAACGUUUG------------UGCAAAA-AGCAAACUC-CCUUCCGCUUU (((((.((((((((.(((((.....))))).))))((((((((((...))--))))))))..)))))))))...(((((------------(......-.))))))..-........... ( -24.60) >DroSec_CAF1 48322 116 + 1 GCAGUCAAAUGGAUAGAUAACACGUUUAUUGAUCCGUUGAAAUAUUGCAC--AUUUCAAUCAAUUUACUGCAACGUUUGCGGCUUCUCUGGCGCAAAA-AGCAAACUC-CUUUCCGCUUU (((((.((((((((.(((((.....))))).))))((((((((.......--))))))))..)))))))))....((((((.(......).))))))(-(((......-......)))). ( -27.80) >DroYak_CAF1 6954 120 + 1 GCAGUCAAAUGGAUAGAUAACACGUUUAUUGAUCCGUUGAAAUAUUGCAUAUAUUUCAAUCAAUUUACUGCAACGUUUUCGGCUCCUCGUGGGUAAAAUGGCAAACUCUCUUUCCCCUUC (((((.((((((((.(((((.....))))).))))((((((((((.....))))))))))..)))))))))..((((((..((.((....)))))))))).................... ( -26.20) >consensus GCAGUCAAAUGGAUAGAUAACACGUUUAUUGAUCCGUUGAAAUAUUGCAU__AUUUCAAUCAAUUUACUGCAACGUUUGCGGCU_CUC__GCGCAAAA_AGCAAACUC_CUUUCCGCUUU (((((.((((((((.(((((.....))))).))))((((((((.........))))))))..)))))))))...((((((....................)))))).............. (-18.55 = -19.22 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:32 2006