
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,128,818 – 20,129,032 |
| Length | 214 |
| Max. P | 0.987488 |

| Location | 20,128,818 – 20,128,937 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
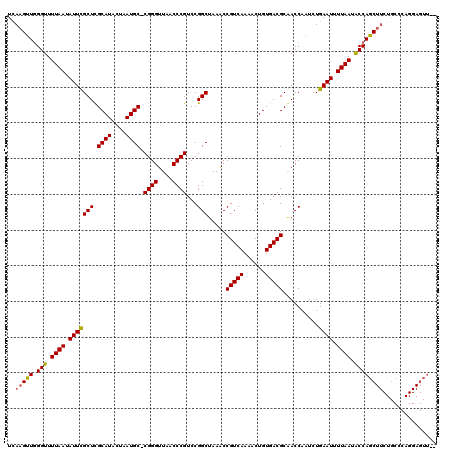
>2L_DroMel_CAF1 20128818 119 - 22407834 UCAAGUUGGGUUUUAAUAUUCGCUCGCAUACUAAUGC-CGGGUUAACCCGUCCGGCUAAACCGUCAAAACUGUGACGCAACCAAUCUGAAUUUUAAUACCAGCUUCUGCCCAGGAGUUUU ........(((.((((.(((((((.((((....))))-((((....))))...))).....(((((......)))))..........)))).)))).)))(((((((....))))))).. ( -30.20) >DroSec_CAF1 40156 117 - 1 UUAAGUUGGGUUUUAAUAUUCGCUCGCAUACUAAUGC-CGGGUUAACCCGUCCGGCUAAACCGUCAAAACUGUGACGCGACCAAUCUGAAUUUUAAAACCAGCUUCUGCCCAGGAGUU-- ........((((((((.(((((.((((........((-((((........))))))......((((......))))))))......))))).))))))))(((((((....)))))))-- ( -34.90) >DroSim_CAF1 3551 117 - 1 UUAAGUUGGGUUUUAAUAUUCGCUCGCAUACUAAUGC-CGGGUUAACCCGUCCGGCUAAACCGUCAAAACUGUGACGCGACCAAUCUGAAUUUUAAUACCAGCUUCUGCCCAGGAGUU-- ........(((.((((.(((((.((((........((-((((........))))))......((((......))))))))......))))).)))).)))(((((((....)))))))-- ( -31.60) >DroEre_CAF1 16821 103 - 1 UCAAGCUAGGUUUUAAUAUUUGCUUGCAUACUAAUGCGCGGGUUAACCCGUCCGGCUAAGCCGUCAAAACUGUGACGCAACCAAUCUGAAUUUUAAUGCCAGC----------------- ....(((.(((.((((.((((((((((((....))))(((((....)))))......))))(((((......)))))..........)))).)))).))))))----------------- ( -27.40) >consensus UCAAGUUGGGUUUUAAUAUUCGCUCGCAUACUAAUGC_CGGGUUAACCCGUCCGGCUAAACCGUCAAAACUGUGACGCAACCAAUCUGAAUUUUAAUACCAGCUUCUGCCCAGGAGUU__ ..(((((.(((.((((.(((((((.((((....)))).((((....))))...))).....(((((......)))))..........)))).)))).))))))))............... (-25.56 = -25.50 + -0.06)


| Location | 20,128,937 – 20,129,032 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
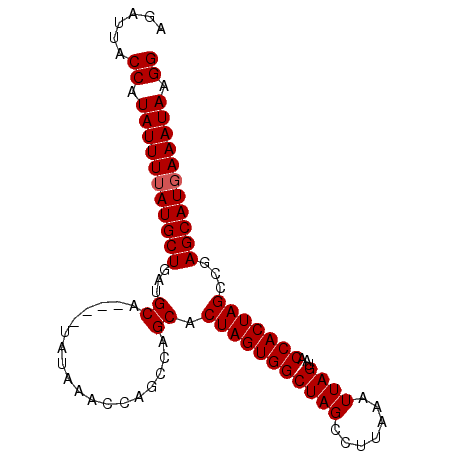
>2L_DroMel_CAF1 20128937 95 + 22407834 AGAUUACCAUAUUUUAUGCUGAUGCAUACAUAUAUACCAGCCAGCACUAGUGGCUAGUCUUAUUUUAGUAACCACUAGCCGAGCAUCAAAUAAGG ......((.(((((.(((((..(((..................)))....((((((((.((((....))))..))))))))))))).))))).)) ( -20.97) >DroSec_CAF1 40273 91 + 1 AGAUUACCAUAUUUUAUGCUGAAGCA----UAUAAACCAGCCAGCACUAGUGGCUAGCCUUAAAUUAGUAACCACUAGCCGAGCAUGAAAUAAGG ......((.(((((((((((...((.----.............)).(((((((((((.......))))...)))))))...))))))))))).)) ( -22.84) >DroSim_CAF1 3668 91 + 1 AGAUUACCAUAUUUUAUGCUGAUGCA----UAUAAGCCAGCCAGCACUAGUGGCUAGCCUUAAAUUAGUAACCACUAGCCGAGCAUGAAAUAAGG ......((.(((((((((((...((.----..(((((.(((((.......))))).).))))...((((....))))))..))))))))))).)) ( -25.00) >consensus AGAUUACCAUAUUUUAUGCUGAUGCA____UAUAAACCAGCCAGCACUAGUGGCUAGCCUUAAAUUAGUAACCACUAGCCGAGCAUGAAAUAAGG ......((.(((((((((((...((..................)).(((((((((((.......))))...)))))))...))))))))))).)) (-21.67 = -22.00 + 0.33)

| Location | 20,128,937 – 20,129,032 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
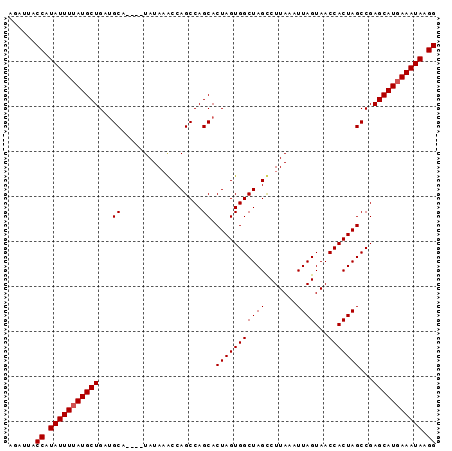
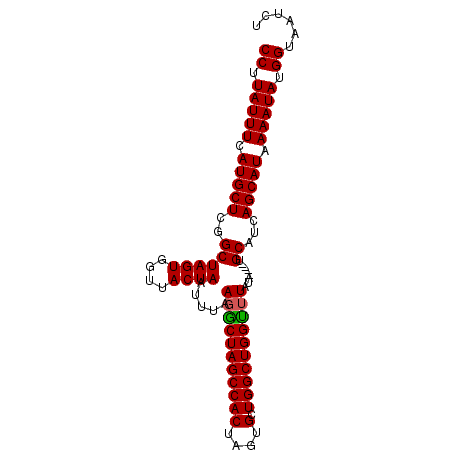
>2L_DroMel_CAF1 20128937 95 - 22407834 CCUUAUUUGAUGCUCGGCUAGUGGUUACUAAAAUAAGACUAGCCACUAGUGCUGGCUGGUAUAUAUGUAUGCAUCAGCAUAAAAUAUGGUAAUCU ((.(((((........(((((((((((((......))..)))))))))))(((((...(((((....))))).)))))...))))).))...... ( -26.80) >DroSec_CAF1 40273 91 - 1 CCUUAUUUCAUGCUCGGCUAGUGGUUACUAAUUUAAGGCUAGCCACUAGUGCUGGCUGGUUUAUA----UGCUUCAGCAUAAAAUAUGGUAAUCU ((.(((((.(((((.(((((((....)))).....(((((((((((....).))))))))))...----.)))..))))).))))).))...... ( -26.20) >DroSim_CAF1 3668 91 - 1 CCUUAUUUCAUGCUCGGCUAGUGGUUACUAAUUUAAGGCUAGCCACUAGUGCUGGCUGGCUUAUA----UGCAUCAGCAUAAAAUAUGGUAAUCU ((.(((((.(((((..((((((....)))).....(((((((((((....).))))))))))...----.))...))))).))))).))...... ( -28.10) >consensus CCUUAUUUCAUGCUCGGCUAGUGGUUACUAAUUUAAGGCUAGCCACUAGUGCUGGCUGGUUUAUA____UGCAUCAGCAUAAAAUAUGGUAAUCU ((.(((((.(((((..((((((....)))).....(((((((((((....).))))))))))........))...))))).))))).))...... (-25.71 = -25.60 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:26 2006