
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,062,911 – 20,063,004 |
| Length | 93 |
| Max. P | 0.697339 |

| Location | 20,062,911 – 20,063,004 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20062911 93 + 22407834 GUAGUUGCUCUUCUCACCCCUU----CGCAGACAGCUGCUAGUUUUCCCUUUGAAUAGGCCUUGACCUUUUUAGAGCGAGACCGACUGGCAACGUAA ...((((((..((.........----.((((....))))..(.((((.(((((((.(((......))).))))))).)))).)))..)))))).... ( -25.90) >DroSec_CAF1 5109 93 + 1 GCAGUUGCUCUUCUCACCCCUU----CGCAGACAGCUGCGAGUUUUCCCUUUGAAUAGGCCUUGACCUUUUCAGAGCGAGAGCGACUGGCAACCUAA .((((((((((((((.....((----(((((....))))))).........((((.(((......))).))))))).)))))))))))......... ( -35.80) >DroSim_CAF1 5075 93 + 1 GCAGUUGCUCUUCUCACCCCUU----CGCAGACAGCUGCGAGUUUUCCCUUUGAAUAGGCCUUGACCUUUUCAGAGCGAGAGCGACUGGCAACGUAA ...((((((..((........(----(((((....))))))((((((.(((((((.(((......))).))))))).))))))))..)))))).... ( -36.10) >DroEre_CAF1 5427 80 + 1 GUAUUUGCUUU--UCACCCUUU----CACAGACAGCUGCGGGUUUUCCUUCUGAAUGGGCCUUGACCUUUUCAGAGCG-----------AAACGUAA ((.(((((...--..((((...----..(((....))).))))......((((((.(((......))).)))))))))-----------)))).... ( -19.20) >DroYak_CAF1 5304 95 + 1 GAAAUUGCUUU--UUACCCGUUUCCCCACAGACAGCUGCGAGUUUUCCUUGUGAAUAGGCCUUGACCUUUUCAGAGCGAGACCAAGUGGAAACGUAA ...........--.....(((((((.(.(((....))).(.(((((.(((.((((.(((......))).))))))).))))))..).)))))))... ( -21.60) >consensus GCAGUUGCUCUUCUCACCCCUU____CGCAGACAGCUGCGAGUUUUCCCUUUGAAUAGGCCUUGACCUUUUCAGAGCGAGACCGACUGGCAACGUAA (((((((.(((..................))))))))))....((((.(((((((.(((......))).))))))).))))................ (-16.07 = -16.39 + 0.32)


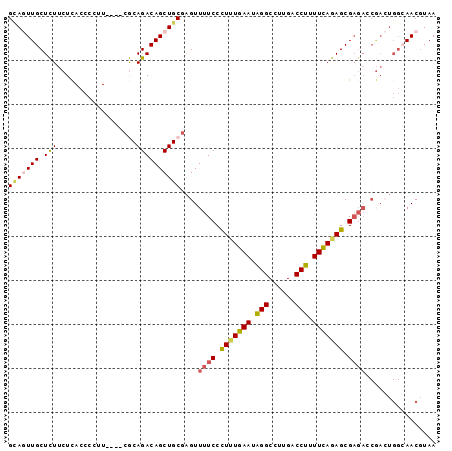
| Location | 20,062,911 – 20,063,004 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -18.06 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20062911 93 - 22407834 UUACGUUGCCAGUCGGUCUCGCUCUAAAAAGGUCAAGGCCUAUUCAAAGGGAAAACUAGCAGCUGUCUGCG----AAGGGGUGAGAAGAGCAACUAC ....(((((...((..(((((((((....((((....)))).(((..((......)).((((....)))))----))))))))))).)))))))... ( -29.80) >DroSec_CAF1 5109 93 - 1 UUAGGUUGCCAGUCGCUCUCGCUCUGAAAAGGUCAAGGCCUAUUCAAAGGGAAAACUCGCAGCUGUCUGCG----AAGGGGUGAGAAGAGCAACUGC ...((((((...((..((((((((((((.((((....)))).)))...........((((((....)))))----).))))))))).)))))))).. ( -35.90) >DroSim_CAF1 5075 93 - 1 UUACGUUGCCAGUCGCUCUCGCUCUGAAAAGGUCAAGGCCUAUUCAAAGGGAAAACUCGCAGCUGUCUGCG----AAGGGGUGAGAAGAGCAACUGC ....(((((...((..((((((((((((.((((....)))).)))...........((((((....)))))----).))))))))).)))))))... ( -34.80) >DroEre_CAF1 5427 80 - 1 UUACGUUU-----------CGCUCUGAAAAGGUCAAGGCCCAUUCAGAAGGAAAACCCGCAGCUGUCUGUG----AAAGGGUGA--AAAGCAAAUAC .....(((-----------(((((((((..(((....)))..)))))).......(((((((....)))))----...))))))--))......... ( -20.40) >DroYak_CAF1 5304 95 - 1 UUACGUUUCCACUUGGUCUCGCUCUGAAAAGGUCAAGGCCUAUUCACAAGGAAAACUCGCAGCUGUCUGUGGGGAAACGGGUAA--AAAGCAAUUUC ...(((((((.(((((.....)..((((.((((....)))).))))))))......((((((....))))))))))))).....--........... ( -25.60) >consensus UUACGUUGCCAGUCGCUCUCGCUCUGAAAAGGUCAAGGCCUAUUCAAAGGGAAAACUCGCAGCUGUCUGCG____AAGGGGUGAGAAGAGCAACUAC ....(((((.......((((((((((((.((((....)))).))))...........(((((....))))).......))))))))...)))))... (-18.06 = -20.00 + 1.94)


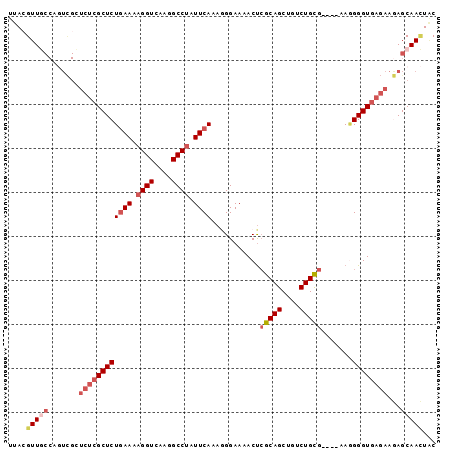
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:12 2006