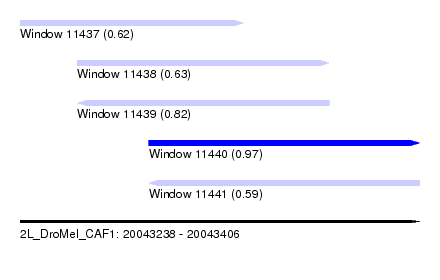
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,043,238 – 20,043,406 |
| Length | 168 |
| Max. P | 0.971695 |

| Location | 20,043,238 – 20,043,332 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.54 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20043238 94 + 22407834 ----GCCAAAUAUUUUGCACAGAGCUCC----------------------UUUGACAGCAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUG ----.............(((((((((..----------------------......))).....((((((((..((((....((((....))))))))..))))))))......)))))) ( -24.10) >DroSec_CAF1 225518 81 + 1 AAAUGCCAACUAUUUUGC---------------------------------------GAAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUACACCCUGUG ..................---------------------------------------...((..((((((((..((((....((((....))))))))..))))))))..))........ ( -18.50) >DroYak_CAF1 219457 120 + 1 ACAAGCCAAUGAUUUCCCGAAGAGCCCCUUUGAUCCAAUUCCCAGUGACUUGGGGCCACAAGUGCCAUUAUAGUUUUACCUUGAAACUACUUUCCAGUCAUAUCAUGGUUUCAUUCUGUG ..((((((.((((....(((((.....)))))............((((((.((((((....).)))....(((((((.....)))))))....))))))))))))))))))......... ( -27.20) >consensus A_A_GCCAAAUAUUUUGC__AGAGC_CC______________________U__G_C_GCAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUG ................................................................((((((((..((((....((((....))))))))..))))))))............ ( -9.95 = -10.73 + 0.78)

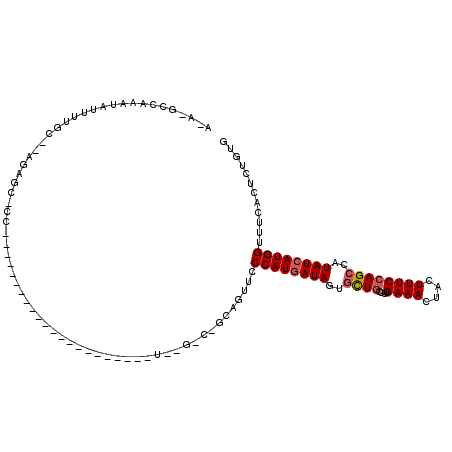

| Location | 20,043,262 – 20,043,368 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -17.85 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20043262 106 + 22407834 ----------UUUGACAGCAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUA ----------....((((.(((..((((((((..((((....((((....))))))))..))))))))....)))))))(((((((((..........)))))).)))........ ( -27.70) >DroSec_CAF1 225536 99 + 1 -----------------GAAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUACACCCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUA -----------------...((..((((((((..((((....((((....))))))))..))))))))..))....((((((((((((..........)))))).).))))).... ( -25.80) >DroEre_CAF1 220453 116 + 1 CCCAGUGGCUUGUGGCCACAGUUCCCAUGAUAGUUUUGCCUUGAAACUACUUUCCUGCCAUAUCAUGGUUUCACUCUGCGGCAUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUA ....(((((.....))))).....((((((((.....((...((((....))))..))..))))))))...(((((((.....))))(((((......)))))))).......... ( -28.40) >DroYak_CAF1 219497 116 + 1 CCCAGUGACUUGGGGCCACAAGUGCCAUUAUAGUUUUACCUUGAAACUACUUUCCAGUCAUAUCAUGGUUUCAUUCUGUGGCGUAGACAGACAAAUAUGUCUGGUGUUUAUAAAUA (((((....)))))((((((((((......(((((((.....)))))))....(((.........)))...)))).)))))).....((((((....))))))............. ( -29.30) >consensus __________UGUGGCCACAGUUCCCAUGAUAGUGCUGCCUUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUA ..............(((((((...((((((((.....((...((((....))))..))..)))))))).......))))))).....(((((......)))))............. (-17.85 = -19.60 + 1.75)


| Location | 20,043,262 – 20,043,368 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -20.88 |
| Energy contribution | -22.12 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20043262 106 - 22407834 UAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGAGUGAAACCAUGAUAUGGCUGGAAAGUAGUUUCAAGGCAGCACUAUCAUGGGAACUGCUGUCAAA---------- .............(((((......)))))........(((((((....((((((((..((((((((....))))....))))..))))))))..))).))))....---------- ( -28.50) >DroSec_CAF1 225536 99 - 1 UAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGGGUGUAACCAUGAUAUGGCUGGAAAGUAGUUUCAAGGCAGCACUAUCAUGGGAACUUC----------------- .........(((((((((......))))((....)).....)))))..((((((((..((((((((....))))....))))..)))))))).......----------------- ( -26.90) >DroEre_CAF1 220453 116 - 1 UAUUUAUAAACACCAGACUUAUUUGUCUGUCUAUGCCGCAGAGUGAAACCAUGAUAUGGCAGGAAAGUAGUUUCAAGGCAAAACUAUCAUGGGAACUGUGGCCACAAGCCACUGGG .............(((((......)))))......((....(((....((((((((..((..((((....))))...)).....))))))))..)))(((((.....))))).)). ( -31.40) >DroYak_CAF1 219497 116 - 1 UAUUUAUAAACACCAGACAUAUUUGUCUGUCUACGCCACAGAAUGAAACCAUGAUAUGACUGGAAAGUAGUUUCAAGGUAAAACUAUAAUGGCACUUGUGGCCCCAAGUCACUGGG .............((((((....)))))).....(((((((......(((.(((.((.(((....))).)).))).)))....(......)....)))))))((((......)))) ( -29.60) >consensus UAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGAGUGAAACCAUGAUAUGGCUGGAAAGUAGUUUCAAGGCAAAACUAUCAUGGGAACUGCGGCCACA__________ .............(((((......))))).....(((((((.......((((((((..((..((((....))))...)).....))))))))...))))))).............. (-20.88 = -22.12 + 1.25)

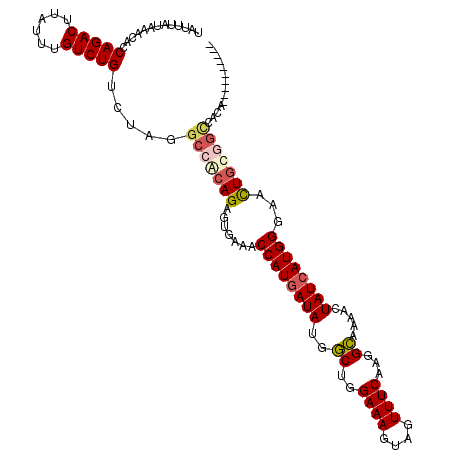

| Location | 20,043,292 – 20,043,406 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20043292 114 + 22407834 UUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUAAACCAUCAUUGCCAUAAUGGCCAUCAUCAGCCAAUGCG .((((((((................))))))))...((((((.....(((((......)))))(.(((((....))))))......)))))).((((........))))..... ( -25.39) >DroSec_CAF1 225559 114 + 1 UUGAAACUACUUUCCAGCCAUAUCAUGGUUACACCCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUAAACCAUCAUUGCCAUAAUGGCCAUCAUCAGCCAAUGCA ...............((((((...))))))......((((((.....(((((......)))))(.(((((....))))))......)))))).((((........))))..... ( -24.40) >DroEre_CAF1 220493 111 + 1 UUGAAACUACUUUCCUGCCAUAUCAUGGUUUCACUCUGCGGCAUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUAAACCAUCAUUGCCAUAAUGGCCAUCAUCAGCCAAU--- .((((((((................))))))))......((((..(((((((......)))))(.(((((....)))))).))..))))....((((........))))..--- ( -24.59) >DroYak_CAF1 219537 114 + 1 UUGAAACUACUUUCCAGUCAUAUCAUGGUUUCAUUCUGUGGCGUAGACAGACAAAUAUGUCUGGUGUUUAUAAAUAAACCAUCAUUGCCAUAAUGGCCAUCAUCAGCCAAUAUA .((((((((................))))))))...(((((((..((((((((....))))))(.(((((....)))))).))..))))))).((((........))))..... ( -27.59) >consensus UUGAAACUACUUUCCAGCCAUAUCAUGGUUUCACUCUGUGGCCUAGACAGACAAAUAAGUCUGGUGUUUAUAAAUAAACCAUCAUUGCCAUAAUGGCCAUCAUCAGCCAAUGCA .((((((((................))))))))...((((((.....(((((......)))))(.(((((....))))))......)))))).((((........))))..... (-23.74 = -24.24 + 0.50)

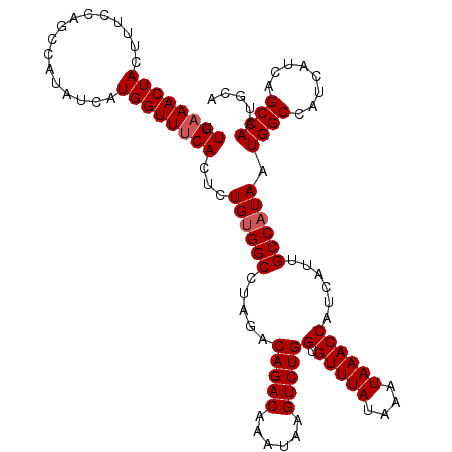
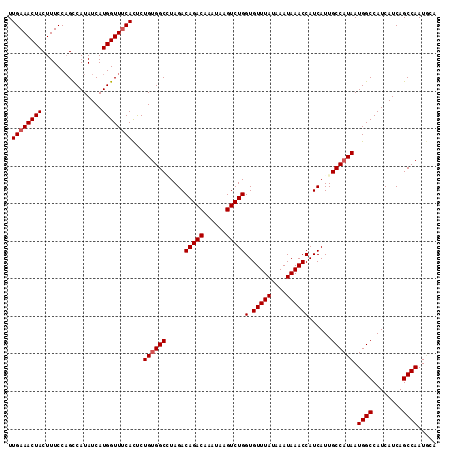
| Location | 20,043,292 – 20,043,406 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -25.95 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20043292 114 - 22407834 CGCAUUGGCUGAUGAUGGCCAUUAUGGCAAUGAUGGUUUAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGAGUGAAACCAUGAUAUGGCUGGAAAGUAGUUUCAA .....(((((.((((.(((((((((....)))))))))...))))......(((((......)))))....))))).....(((((((((...)))(((....))).)))))). ( -29.80) >DroSec_CAF1 225559 114 - 1 UGCAUUGGCUGAUGAUGGCCAUUAUGGCAAUGAUGGUUUAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGGGUGUAACCAUGAUAUGGCUGGAAAGUAGUUUCAA ((((((((((.((((.(((((((((....)))))))))...))))......(((((......)))))....)))))....)))))....(((.((.(((....))).)).))). ( -31.00) >DroEre_CAF1 220493 111 - 1 ---AUUGGCUGAUGAUGGCCAUUAUGGCAAUGAUGGUUUAUUUAUAAACACCAGACUUAUUUGUCUGUCUAUGCCGCAGAGUGAAACCAUGAUAUGGCAGGAAAGUAGUUUCAA ---...(((((((((.(((((((((....)))))))))...))))......(((((......)))))(((.(((((.(..(((....)))..).))))))))...))))).... ( -26.90) >DroYak_CAF1 219537 114 - 1 UAUAUUGGCUGAUGAUGGCCAUUAUGGCAAUGAUGGUUUAUUUAUAAACACCAGACAUAUUUGUCUGUCUACGCCACAGAAUGAAACCAUGAUAUGACUGGAAAGUAGUUUCAA .....((((..((((.(((((((((....)))))))))...))))......((((((....)))))).....)))).....(((((((((...)))(((....))).)))))). ( -29.30) >consensus UGCAUUGGCUGAUGAUGGCCAUUAUGGCAAUGAUGGUUUAUUUAUAAACACCAGACUUAUUUGUCUGUCUAGGCCACAGAGUGAAACCAUGAUAUGGCUGGAAAGUAGUUUCAA .....((((..((((.(((((((((....)))))))))...))))......(((((......))))).....)))).....(((((((((...)))(((....))).)))))). (-25.95 = -26.07 + 0.12)


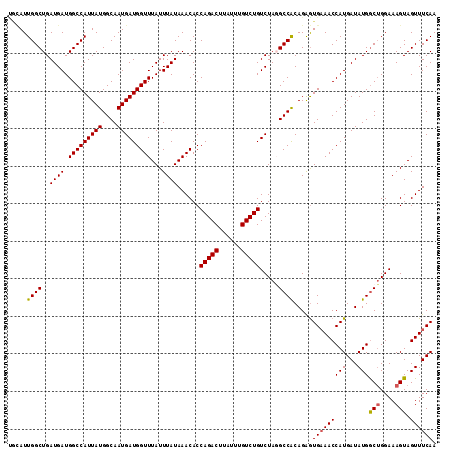
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:04 2006