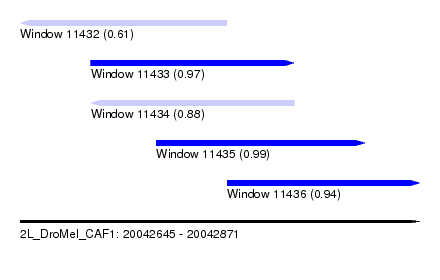
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,042,645 – 20,042,871 |
| Length | 226 |
| Max. P | 0.988784 |

| Location | 20,042,645 – 20,042,762 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.47 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20042645 117 - 22407834 UGGGUUGAGGGUUGAGGGUUUCGGGGUUUUCUCCGGAAAACUGUUCACUUCAGUUCUUGCCACUCAGUGGUGGCAAGCGUGAAAUUGAAAUUUAUGCUCUAAUUACAAACGAAUGCC .(..(((((((...((..((((((((....))))))))..))...).))))))..)((((((((....))))))))(((((((.......))))))).................... ( -37.80) >DroSec_CAF1 224929 107 - 1 UGGGUCG-------AGGGUUUCGGGGUUUUCCCCGGAAAACUCG---GAAAACUGUUUGCUACUCAGUGGUGGCAAGCGUGAAAUUGAAAUUUAUGCUCUAAUUACAAACGAAUGCC .((((((-------((..((((((((....))))))))..))))---.......((((((((((....)))))))))).................)))).................. ( -36.80) >DroEre_CAF1 219805 108 - 1 UGGGUCG-------AGGGCUACGGGGUUUUCCCCGGAAAACUGUUCACUUCAG--UUCGCUACUCAGUGGUGGCAAGCGUGAAAUUGAAAUUUAUGCUCUAAUUACAAAUGAAUGCC ..(((.(-------(((....(((((....)))))(((.....))).)))).(--(((((((((....)))))).((((((((.......))))))))............))))))) ( -30.30) >consensus UGGGUCG_______AGGGUUUCGGGGUUUUCCCCGGAAAACUGUUCACUUCAGU_UUUGCUACUCAGUGGUGGCAAGCGUGAAAUUGAAAUUUAUGCUCUAAUUACAAACGAAUGCC .((.(((.......((..((((((((....))))))))..))................((((((....)))))).((((((((.......))))))))...........)))...)) (-25.68 = -25.47 + -0.21)

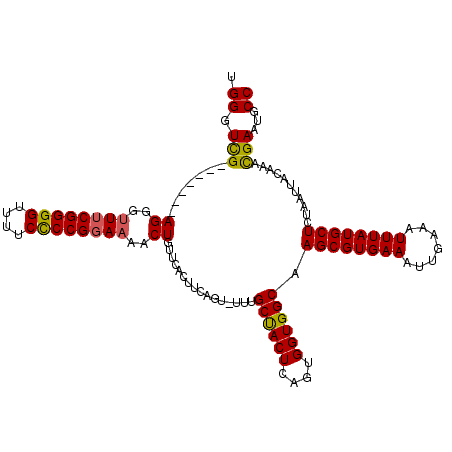
| Location | 20,042,685 – 20,042,800 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.63 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20042685 115 + 22407834 CUUGCCACCACUGAGUGGCAAGAACUGAAGUGAACAGUUUUCCGGAGAAAACCCCGAAACCCUCAACCCUCAACCCACAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAUG ((((((((......)))))))).....(((((....((((((....)))))).....................(((((((......)))))))...........)))))...... ( -32.80) >DroSec_CAF1 224969 105 + 1 CUUGCCACCACUGAGUAGCAAACAGUUUUC---CGAGUUUUCCGGGGAAAACCCCGAAACCCU-------CGACCCACAAGCCCCAUUGUGGGGCUACAUUUAACACUUGGCAUG ..(((((.....(((((((...........---((((..((.(((((....))))).))..))-------)).(((((((......)))))))))))).)).......))))).. ( -39.20) >DroEre_CAF1 219845 106 + 1 CUUGCCACCACUGAGUAGCGAA--CUGAAGUGAACAGUUUUCCGGGGAAAACCCCGUAGCCCU-------CGACCCACAAGCCCCAAAGUUGGGCAACAUUUAACACUUGGCAUG ..(((((....((((..(((((--(((.......))))))..(((((....)))))..)).))-------))........((((.......)))).............))))).. ( -30.50) >consensus CUUGCCACCACUGAGUAGCAAA_ACUGAAGUGAACAGUUUUCCGGGGAAAACCCCGAAACCCU_______CGACCCACAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAUG ..(((((....(((((....................((((..(((((....)))))))))....................(((((.....)))))...))))).....))))).. (-22.85 = -23.63 + 0.78)

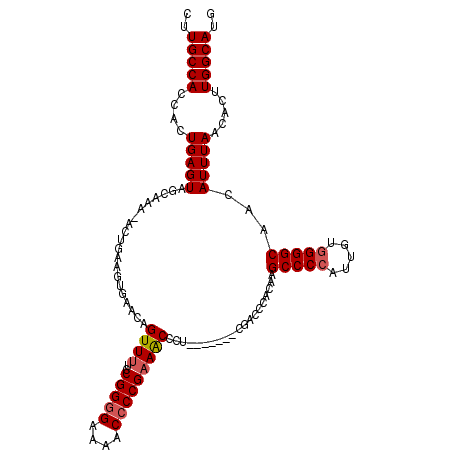
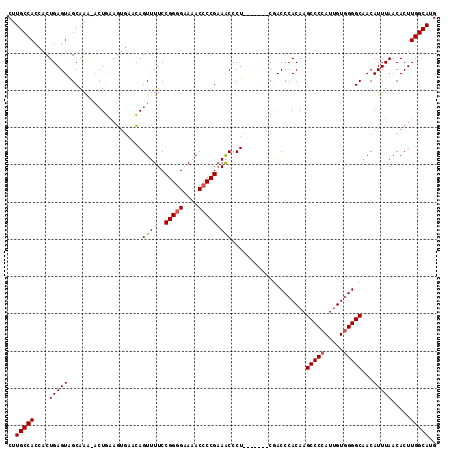
| Location | 20,042,685 – 20,042,800 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -40.61 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20042685 115 - 22407834 CAUGCCAAGUGUUAAAUGUUGCCCCACAAUGGGGCUUGUGGGUUGAGGGUUGAGGGUUUCGGGGUUUUCUCCGGAAAACUGUUCACUUCAGUUCUUGCCACUCAGUGGUGGCAAG ..((((..............(((((.....)))))....(..(((((((...((..((((((((....))))))))..))...).))))))..)..(((((...))))))))).. ( -43.30) >DroSec_CAF1 224969 105 - 1 CAUGCCAAGUGUUAAAUGUAGCCCCACAAUGGGGCUUGUGGGUCG-------AGGGUUUCGGGGUUUUCCCCGGAAAACUCG---GAAAACUGUUUGCUACUCAGUGGUGGCAAG ..(((((..........((((((((((((......)))))))(((-------((..((((((((....))))))))..))))---)..........))))).......))))).. ( -44.83) >DroEre_CAF1 219845 106 - 1 CAUGCCAAGUGUUAAAUGUUGCCCAACUUUGGGGCUUGUGGGUCG-------AGGGCUACGGGGUUUUCCCCGGAAAACUGUUCACUUCAG--UUCGCUACUCAGUGGUGGCAAG ..(((((.............((((.......))))...(((((.(-------.(((((.(((((....)))))(((.....))).....))--))).).)))))....))))).. ( -33.70) >consensus CAUGCCAAGUGUUAAAUGUUGCCCCACAAUGGGGCUUGUGGGUCG_______AGGGUUUCGGGGUUUUCCCCGGAAAACUGUUCACUUCAGU_UUUGCUACUCAGUGGUGGCAAG ..((((..............(((((.....))))).................((..((((((((....))))))))..))................(((((...))))))))).. (-29.41 = -29.97 + 0.56)

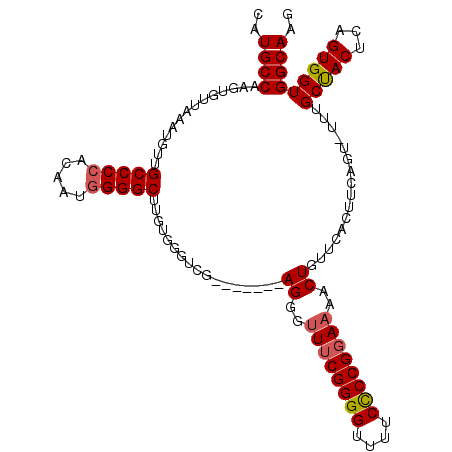
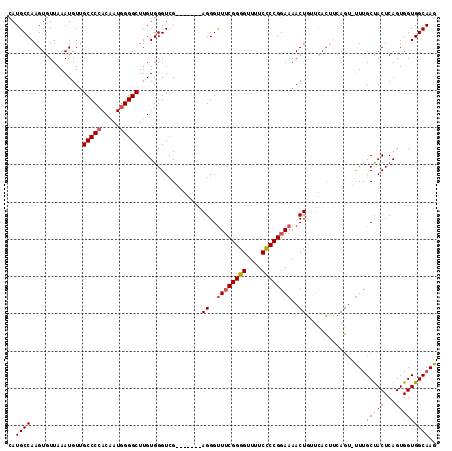
| Location | 20,042,722 – 20,042,840 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -33.43 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20042722 118 + 22407834 UUUUCCGGAGAAAACCCCGAAACCCUCAACCCUCAACCCACAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAUGGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCCACCA .....(((........))).................(((((((......)))))))...............(((((((((.((((.....)))))))(((.....))).))))))... ( -33.30) >DroSec_CAF1 225003 111 + 1 UUUUCCGGGGAAAACCCCGAAACCCU-------CGACCCACAAGCCCCAUUGUGGGGCUACAUUUAACACUUGGCAUGGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCCACCA .....(((((....))))).......-------.........((((((.....))))))............(((((((((.((((.....)))))))(((.....))).))))))... ( -40.20) >DroEre_CAF1 219880 108 + 1 UUUUCCGGGGAAAACCCCGUAGCCCU-------CGACCCACAAGCCCCAAAGUUGGGCAACAUUUAACACUUGGCAUGGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCC---A .....(((((....))))).......-------..........((((.......)))).............(((((((((.((((.....)))))))(((.....))).)))))---) ( -33.90) >consensus UUUUCCGGGGAAAACCCCGAAACCCU_______CGACCCACAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAUGGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCCACCA .....(((((....)))))........................(((((.....)))))..............((((((((.((((.....)))))))(((.....))).))))).... (-33.43 = -34.10 + 0.67)

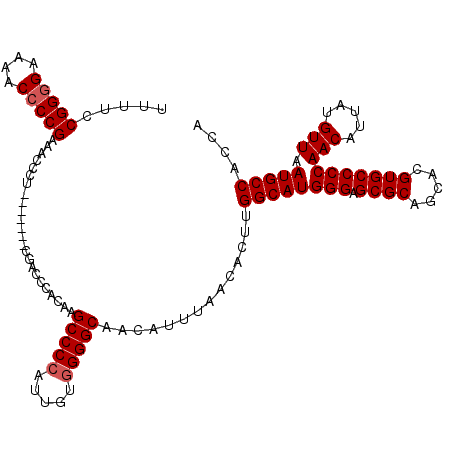
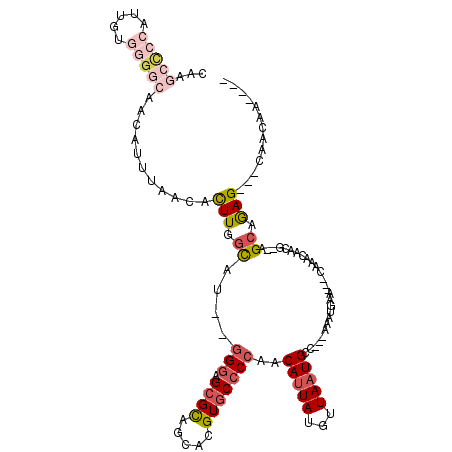
| Location | 20,042,762 – 20,042,871 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -9.92 |
| Energy contribution | -11.88 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20042762 109 + 22407834 CAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAU--GGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCCACCAAAUGAACAACAAACAACG--AGCGGAG---CAACAA---- ...(((((.....)))))..(((((......((((((--(((.((((.....)))))))(((.....))).))))))..)))))..............--.......---......---- ( -31.00) >DroSec_CAF1 225036 109 + 1 CAAGCCCCAUUGUGGGGCUACAUUUAACACUUGGCAU--GGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCCACCAAAUUAACAACAAACAACG--AGCGGAG---CAACAA---- ..((((((.....))))))..........(((.((.(--(((.((((.....))))))))......(((((((........)))))))..........--.)).)))---......---- ( -35.00) >DroEre_CAF1 219913 103 + 1 CAAGCCCCAAAGUUGGGCAACAUUUAACACUUGGCAU--GGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCC---AAGUGAA---CAAACAACG--AGCAGAG---CAACAA---- ...((((.......)))).........((((((((((--(((.((((.....)))))))(((.....))).)))))---)))))..---.........--.......---......---- ( -34.40) >DroYak_CAF1 218957 96 + 1 -------CAUCGUUGGGCAACAUUUAACACUUGGCAU--GGGAGCGUAGCACGUGCCCCAACAUUAUGGUACUGCG---AAAUGAA---CAAACAACG--AGCAAAG---UAACAA---- -------..(((((((....)......((.((.((((--(((.(((.....)))..))))((......))..))).---)).))..---....)))))--)......---......---- ( -21.60) >DroAna_CAF1 234988 109 + 1 -----UUCAUUGUGUGGCAACAUUUAGAAUUUAAUACUGAGAAGAGUAGCACGUGCCCAGACAUUAUGUUAAUGCC---AAAUGAA---CAAACAACAGAGGCAGAGGCGCCACCAGGAG -----.((.(((.(((((................((((......))))......((((.(((.....)))..((((---...((..---.......))..))))).))))))))))))). ( -23.00) >consensus CAAGCCCCAUUGUGGGGCAACAUUUAACACUUGGCAU__GGGAGCGCAGCACGUGCCCCAACAUUAUGUUAAUGCC___AAAUGAA___CAAACAACG__AGCAGAG___CAACAA____ ...(((((.....)))))...........(((.((....(((.((((.....)))))))..(((((...)))))...........................)).)))............. ( -9.92 = -11.88 + 1.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:00 2006