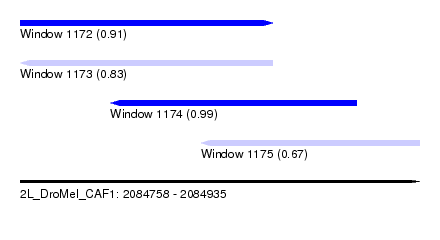
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,084,758 – 2,084,935 |
| Length | 177 |
| Max. P | 0.992479 |

| Location | 2,084,758 – 2,084,870 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -26.21 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2084758 112 + 22407834 AUAAAACUUUGCCGCCUCGAUUGAUUGAUGUGGCCUGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAACCAACCUGUAAGCC--------AGAGUCCUUUGCAAAGGACCUCCUGGCCAAG ......(((((..(((((.......((((...(((((((....))))))).)))))))))))))).............(((--------((.((((((....))))))...))))).... ( -42.01) >DroSec_CAF1 101889 119 + 1 AUAAAACUUUGCCGCCACGAUUGAUUGAUGUGGCCUGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAAACAACCUGGAAGCCAAGGAGCUAGAGUCCU-AGCGAAGGACCUCCUGGCCAAG ......(((.(((............((((...(((((((....))))))).))))((((((..(....)..(((((...)).))).(((((....))-)))...)).))))..))).))) ( -43.20) >DroSim_CAF1 101700 119 + 1 AUAAAACUUUGCCGCCUCGAUUGAUUGAUGUGGCCUGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAAACAACCUGGAAGCCAAGGAGCCAGAGUCCU-GGCGAAGGACCUCCUGGCCAAG ......(((.((((((((.......((((...(((((((....))))))).)))))))))..((((......((((...(....)..)))).(((((-.....))))).))))))).))) ( -47.01) >DroEre_CAF1 104398 99 + 1 AUAAAACUUUGCCGCCUCGAUUGAUUGAUGUGGCCAGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAAGCGACCUGGAAGCAAAGGA---------------------GCUCCUGGCCAAG .....((((.(((((.((((....)))).)))))..(((....))))))).......(((((.(((.((..(((........))).---------------------))))))))))... ( -35.30) >DroYak_CAF1 105991 100 + 1 AUAAAACUUUGCCGCCUCGAUUGAUUGAUGUGGCCUGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAAACGACCUGGAAGCCAGGGU--------C------------CCUCUUGGCCAAG ..........(((((.((((....)))).)))))(((((....))))).........((((((((......(((((...)))))..--------.------------..))))))))... ( -39.70) >DroAna_CAF1 101244 99 + 1 AUAAAAGUUUGCCGCCUCGAUUGAUUGAUGUGGCCAGGCAUUUGCCAGGUAAUCAAUGGCCAAGGAUGCC-CGUCGAUGCCA--------------------AAGGACCUUCUGGCCAUG ......(((((((((.((((....)))).)))).)))))....((((((...((..((((....(((...-.)))...))))--------------------...))...)))))).... ( -29.60) >consensus AUAAAACUUUGCCGCCUCGAUUGAUUGAUGUGGCCUGGCAUUUGCCAGGUAAUCAGAGGCCAAGGAAACAACCUGGAAGCCAAGGA________C_______AAGGACCUCCUGGCCAAG ..........(((((.((((....)))).)))))(((((....))))).........(((((.(((...........................................))))))))... (-26.21 = -26.43 + 0.22)

| Location | 2,084,758 – 2,084,870 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -22.76 |
| Energy contribution | -24.95 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2084758 112 - 22407834 CUUGGCCAGGAGGUCCUUUGCAAAGGACUCU--------GGCUUACAGGUUGGUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCAGGCCACAUCAAUCAAUCGAGGCGGCAAAGUUUUAU ...((((....))))((((((.(((((..(.--------.(((....)))..).))))).(((((((((..((((((....))))))....)))).......))))).))))))...... ( -41.71) >DroSec_CAF1 101889 119 - 1 CUUGGCCAGGAGGUCCUUCGCU-AGGACUCUAGCUCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCAGGCCACAUCAAUCAAUCGUGGCGGCAAAGUUUUAU ...((((((((((((....(((-((....))))).....)))))))(((......)))))))).........(((((....)))))(((((...........)))))............. ( -42.80) >DroSim_CAF1 101700 119 - 1 CUUGGCCAGGAGGUCCUUCGCC-AGGACUCUGGCUCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCAGGCCACAUCAAUCAAUCGAGGCGGCAAAGUUUUAU (((.(((.(((((((....(((-((....))))).....)))))))(((......)))..(((((((((..((((((....))))))....)))).......)))))))).)))...... ( -45.61) >DroEre_CAF1 104398 99 - 1 CUUGGCCAGGAGC---------------------UCCUUUGCUUCCAGGUCGCUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCUGGCCACAUCAAUCAAUCGAGGCGGCAAAGUUUUAU ...((((.(((((---------------------......)).))).))))(((.((((((....((((..((.(((....))).))....)))).....)))))).))).......... ( -31.30) >DroYak_CAF1 105991 100 - 1 CUUGGCCAAGAGG------------G--------ACCCUGGCUUCCAGGUCGUUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCAGGCCACAUCAAUCAAUCGAGGCGGCAAAGUUUUAU ...(((((((.(.------------.--------(((((((...)))))..))..))))))))..((((..((((((....))))))....)))).......(((((......))))).. ( -38.70) >DroAna_CAF1 101244 99 - 1 CAUGGCCAGAAGGUCCUU--------------------UGGCAUCGACG-GGCAUCCUUGGCCAUUGAUUACCUGGCAAAUGCCUGGCCACAUCAAUCAAUCGAGGCGGCAAACUUUUAU .((((((((...((((.(--------------------((....))).)-)))....)))))))).........(((....)))..(((.(.((........)).).))).......... ( -28.10) >consensus CUUGGCCAGGAGGUCCUU_______G________UCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAAUGCCAGGCCACAUCAAUCAAUCGAGGCGGCAAAGUUUUAU ....(((.(((((((........................)))))))..............(((((((((..((((((....))))))....)))).......)))))))).......... (-22.76 = -24.95 + 2.20)

| Location | 2,084,798 – 2,084,907 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.92 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2084798 109 - 22407834 UA--UAAAUAUCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUUGCAAAGGACUCU--------GGCUUACAGGUUGGUUCCUUGGCCUCUGAUUACCUGGCAAA ..--.......((-(((..(((((((((.(((.....))).)))))))))((((((((....)))))).))--------((((...(((......))).))))........))))).... ( -41.00) >DroSec_CAF1 101929 118 - 1 UAUAUAAAUACCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUCGCU-AGGACUCUAGCUCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAA ...........((-((((((((((((((.(((.....))).)))))))))(((((((.....-))))).))...)))))))...((((((.(((............))).)))))).... ( -43.00) >DroSim_CAF1 101740 118 - 1 UAUAUAAAUAUCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUCGCC-AGGACUCUGGCUCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAA ...........((-(((....(((((((.(((.....))).)))))))(((((((....(((-((....)))))..(((((...)))))..........))))))).....))))).... ( -43.60) >DroEre_CAF1 104438 97 - 1 UA--UAAAUAUCCAAGGGACUUGGUCGACUUGUUUCUCAACUUGGCCAGGAGC---------------------UCCUUUGCUUCCAGGUCGCUUCCUUGGCCUCUGAUUACCUGGCAAA ..--........((((((((((((((((.(((.....))).))))))))).((---------------------.(((........)))..)))))))))(((...........)))... ( -28.70) >DroYak_CAF1 106031 98 - 1 UA--UAAAUAUCCCAGGGACUUGGUUGACUUGUUUCUUAACUUGGCCAAGAGG------------G--------ACCCUGGCUUCCAGGUCGUUUCCUUGGCCUCUGAUUACCUGGCAAA ..--........(((((.....((((((........)))))).(((((((.(.------------.--------(((((((...)))))..))..))))))))........))))).... ( -34.40) >DroAna_CAF1 101284 94 - 1 ----UAAAUAUCC-AGAGACGUAUUCAAUUUGUUUCCCAACAUGGCCAGAAGGUCCUU--------------------UGGCAUCGACG-GGCAUCCUUGGCCAUUGAUUACCUGGCAAA ----.......((-((....(((.((((..((((....))))(((((((...((((.(--------------------((....))).)-)))....))))))))))).))))))).... ( -25.30) >consensus UA__UAAAUAUCC_AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUU_______G________UCCUUGGCUUCCAGGUUGUUUCCUUGGCCUCUGAUUACCUGGCAAA ...........((.(((..(((((((((.(((.....))).))))))))).............................((((...(((......))).))))........))))).... (-21.24 = -21.97 + 0.72)
| Location | 2,084,838 – 2,084,935 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.79 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -9.82 |
| Energy contribution | -11.72 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2084838 97 - 22407834 GUUUAAAGGAAUAUUGAGAGAAAAA--------UAAUA--UAAAUAUCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUUGCAAAGGACUCU--------G .......(((.((((..........--------.....--..)))))))-.....(((((((((.(((.....))).)))))))))((((((((....)))))).))--------. ( -27.33) >DroPse_CAF1 136828 100 - 1 AAUGAAAGGAAC---GAAAGGAAUCCUUGAGGA-AAAA--UAAAUAUCU-AGCGACGUUUCAAAUUUGUUUCUCAAGUUAGUU-GGAGCUCCUUUAG-------CCUUGGUC-CAG .....((((...---.((((((.(((....)))-....--......(((-(((((....)).((((((.....)))))).)))-)))..))))))..-------))))....-... ( -20.90) >DroSec_CAF1 101969 106 - 1 GUUUAAAGGAAUAUUGAAAGAAAAA--------UAAUAUAUAAAUACCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUCGCU-AGGACUCUAGCUCCUUG .....(((((...............--------................-((((((((((((((.(((.....))).)))))))...))))))).(((-((....)))))))))). ( -32.10) >DroSim_CAF1 101780 106 - 1 GUUUAAAGGAAUAUUGAAAGAAAAA--------UAAUAUAUAAAUAUCC-AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUUCGCC-AGGACUCUGGCUCCUUG .....(((((...............--------................-((((((((((((((.(((.....))).)))))))...))))))).(((-((....)))))))))). ( -34.10) >DroYak_CAF1 106071 86 - 1 GUUUAAAGGAAUAUUGAAAGAAAAA--------UAAUA--UAAAUAUCCCAGGGACUUGGUUGACUUGUUUCUUAACUUGGCCAAGAGG------------G--------ACCCUG .......(((.((((..........--------.....--..)))))))(((((.((((((..(.(((.....))).)..))))))...------------.--------.))))) ( -18.23) >DroAna_CAF1 101323 80 - 1 GUUUAAAGGAAUCUUUUAAGAAAAA---------------UAAAUAUCC-AGAGACGUAUUCAAUUUGUUUCCCAACAUGGCCAGAAGGUCCUU--------------------UG ...(((((((.((((((........---------------.........-.((((((.........))))))((.....))..)))))))))))--------------------)) ( -14.00) >consensus GUUUAAAGGAAUAUUGAAAGAAAAA________UAAUA__UAAAUAUCC_AGGGACUUGGCCGACUUGUUUCGCAACUUGGCCAGGAGGUCCUU_GC____G__UCU___UC__UG .....(((((....................................((.....))(((((((((.(((.....))).)))))))))...)))))...................... ( -9.82 = -11.72 + 1.89)

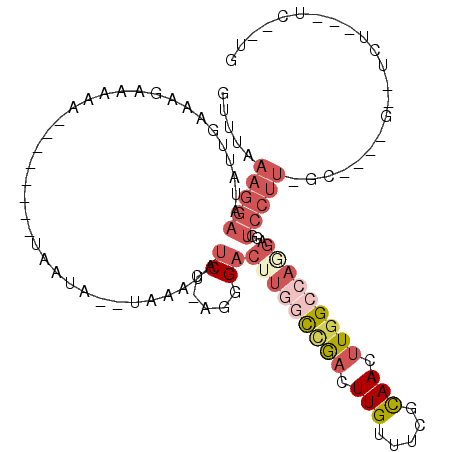
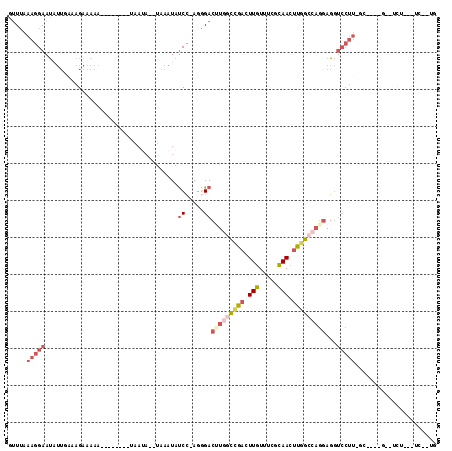
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:44 2006