
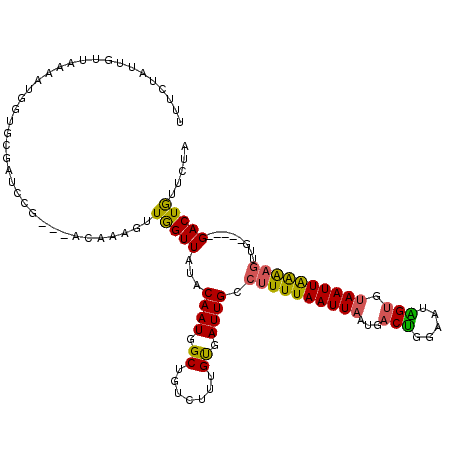
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,039,209 – 20,039,346 |
| Length | 137 |
| Max. P | 0.998669 |

| Location | 20,039,209 – 20,039,321 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.22 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.36 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20039209 112 + 22407834 UUUCUAUUGUUAAAAUGGUGCGAUCCG---ACAAAGUUGGUUAUACAAUGGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUUG-----GACUAUUCCA ..((....((((((..((.((((((..---((((((.(((((((...))))))).))))))))))))))))))))....))..((((((((.((((.....))))-----.)))))))). ( -32.00) >DroSec_CAF1 221539 112 + 1 UUUCUAUUGUUCAAAUGGUGCGAUCCG---ACAAAGUUGGUUAUACAAUAGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUGGUGUAAUUAAAAGUUG-----GACUGUUCUA ........((((((.....((((((..---((((((.(((((((...))))))).))))))))))))((((((((((...(((.....))).)))))))))))))-----)))....... ( -28.00) >DroEre_CAF1 216493 100 + 1 UUUCUAUUGUU------------GCCG---ACAAAGUUAGUUACACAAUUGCAGUCUUUGUGAUUGCCUUUUAAUUAACGACUGCAAUAGUGUAAUUAAAAGUUG-----GACUGUUCUA ...........------------.(((---((....((((((((((.(((((((((....((((((.....))))))..))))))))).))))))))))..))))-----)......... ( -32.30) >DroYak_CAF1 215254 112 + 1 UUUCUAUUGUUUAAAUGGUAUGCUCCU---ACAAAGUUAGUUACACAAUUGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUGG-----GACUGUUCUA ...(((((.....))))).....((((---((....((((((((((.(((.(.(((....((((((.....))))))..))).).))).))))))))))..))))-----))........ ( -27.10) >DroAna_CAF1 232136 120 + 1 UUGGCAUUGUCAAAUUGAGUUUUUCCGUAUAUGAAGGUGGUUUUUCAAUGGCUUUCAUUGCAAUUGUAGUUUAAUUAAUGCCCGGAAUGGAGCAAUUGGGGCGUGGUAAUGACUAUUGGA (((((...)))))..........((((((.((((((((.(((....))).)))))))))))....((((((......((((((.((.((...)).)).))))))......)))))).))) ( -31.20) >consensus UUUCUAUUGUUAAAAUGGUGCGAUCCG___ACAAAGUUGGUUAUACAAUGGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUUG_____GACUGUUCUA .....................................(((((...((((.((.......)).)))).((((((((((...(((.....))).))))))))))........)))))..... (-10.94 = -10.22 + -0.72)


| Location | 20,039,246 – 20,039,346 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.44 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20039246 100 + 22407834 UUAUACAAUGGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUUG-----GACUAUUCCAAUAC-------GACGAGUCAUGAAAACUGAAA-------- .......(((((((((...((.((((.........)))).))(((((((((.((((.....))))-----.)))))))))....-------))).))))))...........-------- ( -24.70) >DroSec_CAF1 221576 100 + 1 UUAUACAAUAGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUGGUGUAAUUAAAAGUUG-----GACUGUUCUAACAG-------GACGAGUCAUAAAAACUGAAA-------- .............((.(((((((((..((((((((((...(((.....))).)))))))))).((-----..((((....))))-------..))))))))))).)).....-------- ( -21.60) >DroEre_CAF1 216518 99 + 1 UUACACAAUUGCAGUCUUUGUGAUUGCCUUUUAAUUAACGACUGCAAUAGUGUAAUUAAAAGUUG-----GACUGUUCUAACGC-------GACGAAUCAUAA-AACUAAAU-------- ((((((.(((((((((....((((((.....))))))..))))))))).))))))......((((-----((....))))))..-------............-........-------- ( -26.40) >DroYak_CAF1 215291 100 + 1 UUACACAAUUGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUGG-----GACUGUUCUAACAC-------GACGGGUCAUAAAAACUAAAU-------- ((((((.(((.(.(((....((((((.....))))))..))).).))).))))))......((((-----..(((((.......-------))))).))))...........-------- ( -18.00) >DroAna_CAF1 232176 120 + 1 UUUUUCAAUGGCUUUCAUUGCAAUUGUAGUUUAAUUAAUGCCCGGAAUGGAGCAAUUGGGGCGUGGUAAUGACUAUUGGAAGAGUAUUGAGUAUGGGUUAUAAAAACAUAAAGGUUUCAA ..(((((((((...((((((((((((.....))))).((((((.((.((...)).)).)))))).))))))))))))))))(((.(((...((((...........))))..)))))).. ( -28.10) >consensus UUAUACAAUGGCUGUCUUUGUGAUUGCCUUUUAAUUAAUGACUGGAAUAGUGUAAUUAAAAGUUG_____GACUGUUCUAACAC_______GACGAGUCAUAAAAACUAAAA________ .............((.(((((((((..((((((((((...(((.....))).))))))))))...........(((....)))............))))))))).))............. ( -9.80 = -9.44 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:54 2006