| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,036,777 – 20,036,887 |
| Length | 110 |
| Max. P | 0.873743 |

| Location | 20,036,777 – 20,036,887 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -19.75 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
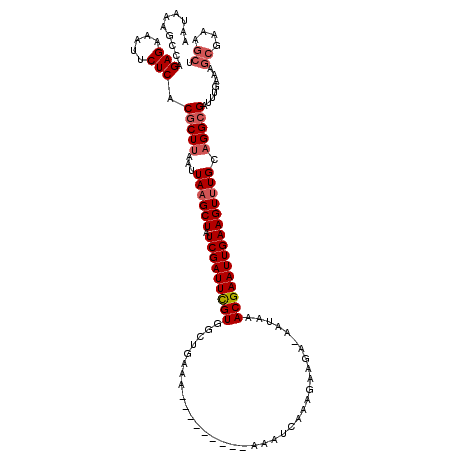
>2L_DroMel_CAF1 20036777 110 + 22407834 AAUAAAACCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUCGUGGCUGAAA---------AAAUCAAAGAAGA-GAUAAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGCU ..........(((.....))).(((((...((((((.(((((((((..((....---------............)-)....))))))))))))))).))))).......(((....))) ( -26.39) >DroSec_CAF1 219046 111 + 1 AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUCGUGGCUGAAAA--------AAAUCAAAGAAGA-GAUGAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGCU ......(((.(((.....))).........((((((.(((((((((.........--------..(((........-)))..)))))))))))))))..)))........(((....))) ( -26.62) >DroSim_CAF1 216400 110 + 1 AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUCGUGGCUGAAA---------AAAUCAAAGAAGA-GAUAAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGAU ......((..(((.....))).(((((...((((((.(((((((((..((....---------............)-)....))))))))))))))).)))))........))....... ( -23.19) >DroEre_CAF1 213789 110 + 1 AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUUGUGGGUGAAA---------AAAUCAAAGAAGA-AAUAAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGCU ......(((.(((.....))).........((((((.(((((((((..((....---------.............-.))..)))))))))))))))..)))........(((....))) ( -25.07) >DroYak_CAF1 212540 110 + 1 AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUUGUGGUUGAAA---------AAAUCAAAGAAGA-AAUAAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGCU ......(((.(((.....))).........((((((.(((((((((((((....---------.)))).....(..-..)..)))))))))))))))..)))........(((....))) ( -25.30) >DroAna_CAF1 229661 120 + 1 AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUUGUGGCUCAAAAAAAAUACAAAAUCAAAGAAGAAAAUAAACGAAUUGAAGUCUGCUGGAGAUUUGAAAGCAAAAACC ..((((.(((((((.........((((....))))..(((((((((....................................)))))))))..))).))))...))))............ ( -17.62) >consensus AAUAAAGCCAGAGAAAUUCUCACGCUUAAUUAAGCUAUCGAUUCGUGGCUGAAA_________AAAUCAAAGAAGA_AAUAAACGAAUUGAAGUUUGCAGGCGAUUUGAAAGCGAAAGCU ..........(((.....))).(((((...((((((.(((((((((....................................))))))))))))))).)))))........((....)). (-19.75 = -20.34 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:51 2006