
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,034,103 – 20,034,212 |
| Length | 109 |
| Max. P | 0.843023 |

| Location | 20,034,103 – 20,034,212 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -16.45 |
| Energy contribution | -17.87 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20034103 109 - 22407834 CGCUUUAAAGGCUUUUAUGCCGCC---AAAAAAAUGGGGGC-AG--CGCUAAUGGGUGGCGGCUG---AAAGUCAAAAGGCACCAGCCCUG-AAUGA-UGUGAUUCAGCUGGCAACUUAA .(((((...((((((((.((((((---(......(((.(..-..--).))).....)))))))))---))))))..))))).(((((..((-(((..-....))))))))))........ ( -39.90) >DroSec_CAF1 216470 112 - 1 CGCUUUAAAGGCUUUUAUGCCCCCCCAAAAAAAAUGGGGGC-AA--CGCUAAUGGGUGGCGGCUG---AAAGUCAAAAGGCAACAGCCCUG-AAUGA-UGUUAUUCAGCUGGCAACUUAA .(((((...((((((((.(((((((((.......)))))).-..--.((((.....)))))))))---))))))..)))))....((((((-((((.-...)))))))..)))....... ( -41.40) >DroSim_CAF1 213916 112 - 1 CGCUUUAAAGGCUUUUAUGCCCCAAAAAAAAAAAAUGGGGC-AA--CGCUAAUGGGUGGCGGCUG---AAAGUCAAAAGGCAACAGCCCUG-AAUGA-UGUGAUUCAGCUGGCAACUUAA .(((((...(((((((((((((((...........))))))-).--(((((.....)))))..))---))))))..)))))....((((((-(((..-....))))))..)))....... ( -38.70) >DroEre_CAF1 211163 112 - 1 CGCUUUAAAGGCUUUUAUACCCCC---GAAAAAAUGGGGGCUAAUGGGCUAAUGGGUGGCGGCGG---AAGGUCAAAAGGCCCCGGCCAUG-AAUGA-UGUGAUUCAGCUGGCAACUUAA ((((.....(((((.....(((((---(......)))))).....))))).......)))).(((---..((((....)))))))((((((-(((..-....)))))..))))....... ( -35.50) >DroYak_CAF1 210003 96 - 1 CGCUUUAAAGGCUUUUAUACCCCC---GAAAAAAAGGGGC--AC--UGCUAAUGGGUGGCGGCGG---AAAGUCAAAAGACUGUGGCCCUG-AAGGA-UGUGAUUCAG------------ .(((.....)))........((((---........))))(--((--(.((...((((.(((((..---..........).)))).))))..-.)).)-.)))......------------ ( -24.80) >DroAna_CAF1 227146 113 - 1 GGUUUUAAGGCCUCUUUCGUACCA---GAAAAAAGUGGACC-AA--CUCUAAUGGGUGGCGGCGGCAGUAAGCCAAAGGACC-UGACUCUGCAAUGGGGGCAAGGCAGUCGGAAAUUUAA ((((((...((((((..(((..((---((.....((.(.((-(.--(((....))))))).))(((.....)))........-....))))..)))..))..)))).....))))))... ( -30.00) >consensus CGCUUUAAAGGCUUUUAUGCCCCC___AAAAAAAUGGGGGC_AA__CGCUAAUGGGUGGCGGCGG___AAAGUCAAAAGGCAACAGCCCUG_AAUGA_UGUGAUUCAGCUGGCAACUUAA .(((.....)))......((((((...........))))))......(((..(((((.(((((........)))....(((....)))...........)).)))))...)))....... (-16.45 = -17.87 + 1.42)

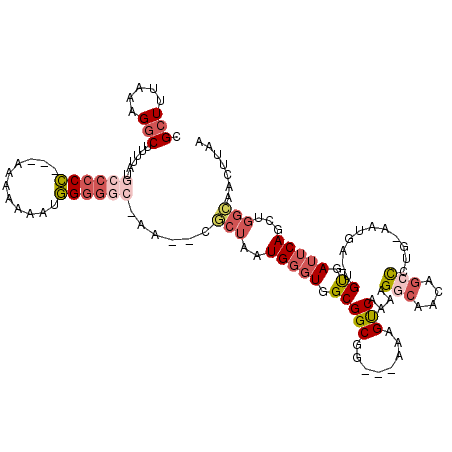
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:50 2006