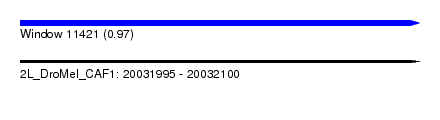
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,031,995 – 20,032,100 |
| Length | 105 |
| Max. P | 0.968691 |

| Location | 20,031,995 – 20,032,100 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -9.01 |
| Energy contribution | -10.23 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031995 105 + 22407834 CCACAUAUCCGUAUCCA-----UAUCCACUGAAU--ACGGAUAUGG---ACA-CCCGGCGUAUGCGUGAUAUCUGCAAC-AGCUCGUAUCUCCAUACU---GCCUUUAAGAUGCCCAUCA ...(((((((((((.((-----.......)).))--))))))))).---...-...((((((((.(.(((((..((...-.))..))))).)))))).---))).....(((....))). ( -34.10) >DroSec_CAF1 214398 105 + 1 GCUCACAU------CCA-----UAUAUACUGAAU--UUAUAUACGUGU-CCAUCCCGGCGUAUGCGUGAUAUCUGCAAC-AGCACUUAUCUCCAUACUGCUGCCUUUAAGAUGCCCAUCA ((((((((------..(-----((((........--.)))))..))))-......(((((((((.(.((((..(((...-.)))..)))).)))))).)))).......)).))...... ( -22.00) >DroSim_CAF1 211728 111 + 1 GCUCAUAUCCGUAUCCA-----UAUAUACUGAAU--UUAUAUACGUGU-CCAUCCCGGCGUAUGCGUGAUAUCUGCAAC-AGCACUUAUCUCCAUACUGCUGCCUUUAAGAUGCCCAUCA ..........(((((..-----((((((......--.)))))).....-......(((((((((.(.((((..(((...-.)))..)))).)))))).)))).......)))))...... ( -22.70) >DroEre_CAF1 208890 100 + 1 CCACAUAU------CCA-----UAUCCAUUGAAU--AGGUUUAUGG---ACA-CCCGGCGUAUGCGUGAUAUCUCCCCCAUGCACUUAUCUUCAUACU---GCCUUUAAGAUGGCCAUCA .......(------(((-----((.((.......--.))..)))))---)..-...(((...((((((..........))))))..((((((.(....---....).))))))))).... ( -21.00) >DroYak_CAF1 207734 99 + 1 CCACAUAU------CCA-----UAUCCAUUAAAA--AAGUUUAUGG---ACA-CCCGGCGUAUGCGUGAUAUCUGCGAC-UGCACUUAUCUCCAUACU---GCCUUUAAGAUGCCCGUCA .......(------(((-----((..(.......--..)..)))))---)..-...((((((((.(.((((..(((...-.)))..)))).)))))).---))).....(((....))). ( -24.80) >DroAna_CAF1 224764 113 + 1 CAACACAGCCUUCGCCUUAUUUUAUCUUCCCAAGGCAAUUUCAUGG--CAUA-CGCUGCGUAUACGCGAUAUUUACAUC-AGCAUUUAU--CCAUUAUGGAG-CUUGAAGAUGCCAAUCA .................................((((.(((((.((--(...-.((((((....)))(((......)))-))).....(--((.....))))-))))))).))))..... ( -24.70) >consensus CCACAUAU______CCA_____UAUCCACUGAAU__AAGUAUAUGG___ACA_CCCGGCGUAUGCGUGAUAUCUGCAAC_AGCACUUAUCUCCAUACU___GCCUUUAAGAUGCCCAUCA ........................................................((((((((.(.((((..(((.....)))..)))).))))))....))).....(((....))). ( -9.01 = -10.23 + 1.22)

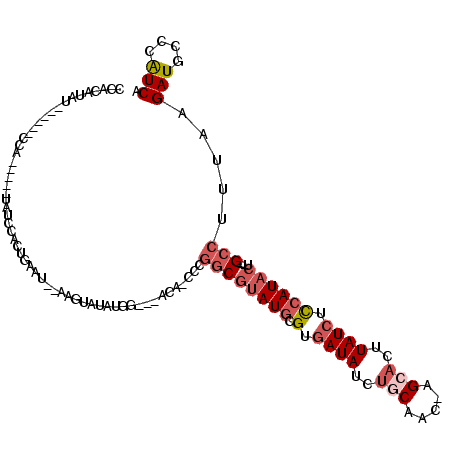

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:46 2006