
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,031,758 – 20,031,891 |
| Length | 133 |
| Max. P | 0.684934 |

| Location | 20,031,758 – 20,031,854 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031758 96 - 22407834 CGGGACGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGAAGCCGCCUCUUUU--------AGGCAGUUGGUCUCGUCUG (((((((((((.........)))))(((....(((((.((..(((((.....)))))..)))))))..((((.....--------)))))))..)))))).... ( -22.70) >DroSec_CAF1 214161 96 - 1 CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGAAGCCGCCUCUUUU--------AGGCAGUUGGUUUCGUCUG .((((((((((.........)))))))))).........(..(((((.....)))))..)((((((((((((.....--------))))....))))))))... ( -29.30) >DroSim_CAF1 211492 96 - 1 CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGAAGCCGCCUCUUUU--------AGGCAGUUGGUCUCGUCUG .((((((((((.........)))))))))).........(..(((((.....)))))..)((((..((((((.....--------))))....))..))))... ( -26.50) >DroEre_CAF1 208647 104 - 1 CAGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUGCUCGGAAUGAACAAGAAGCCGGCUCUUUUGCCUAAACAGACAGUUGGUCUCGUCUG .((((((((((.........))))))))))........((((((...((.(((.............))))).....))))))..(((((((.....)).))))) ( -29.92) >DroYak_CAF1 207498 96 - 1 CGGGGCGUUAUUUAAUUUAUAUAACGUUCUUAUUUUGAUGGGCAUUUUCGCUGAAUGAACACGAAGCUGCCUCUUUU--------AGGCAGUUGGUCUCGUCUG .((((((((((.........))))))))))......(((((((....(((.((......)))))((((((((.....--------)))))))).))).)))).. ( -28.30) >consensus CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGAAGCCGCCUCUUUU________AGGCAGUUGGUCUCGUCUG .((((((((((.........))))))))))......(((((((((((.....))))).......(((.((((.............)))).)))...)))))).. (-20.96 = -20.92 + -0.04)

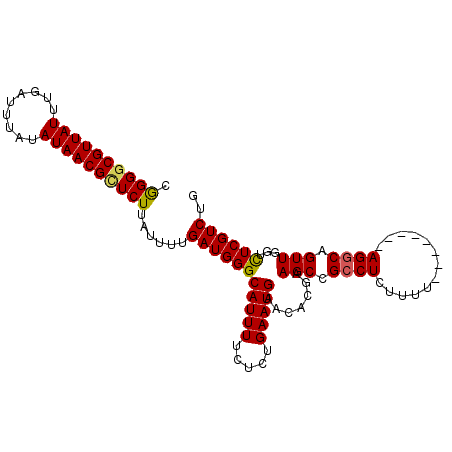
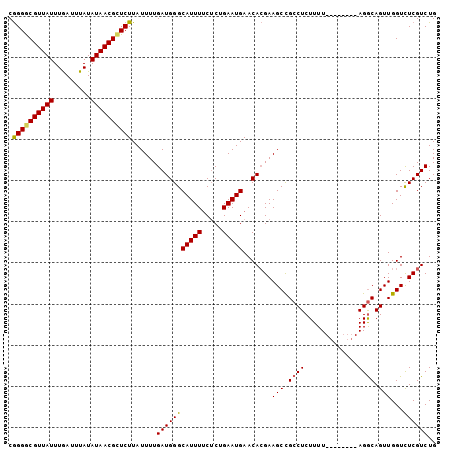
| Location | 20,031,790 – 20,031,891 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031790 101 - 22407834 CUCGUUAAGUGCCUGCGAAAAUGAUUUU---CAUUUAUGC----------------CGGGACGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGA .((((((((((((((((.(((((.....---))))).)))----------------.(((.((((((.........)))))))))..........))))))))......)))))...... ( -23.10) >DroSec_CAF1 214193 101 - 1 CUCGUUAAGUGCCUGCGAAAAUGAUUUU---CAUUUAUGC----------------CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGA .((((((((((((((((.(((((.....---))))).)))----------------.((((((((((.........)))))))))).........))))))))......)))))...... ( -27.40) >DroSim_CAF1 211524 100 - 1 CUCGUUAAGUGCCGGC-AAAAUGAUUUU---CAUUUAUGC----------------CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGA (((((((((..(((((-((((((.....---))))).)))----------------)))((((((((.........)))))))).....)))))))))(((((.....)))))....... ( -30.60) >DroEre_CAF1 208687 100 - 1 CUCGUUAAGUGCCUGUG-AAAUGAUUUU---CAUUUAUGC----------------CAGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUGCUCGGAAUGAACAAGA .(((.(((((((((((.-(((((.....---)))))....----------------.((((((((((.........)))))))))).......)))))))))))..)))........... ( -27.50) >DroYak_CAF1 207530 100 - 1 CUCGUUAAGUGUCUGUG-CAAUGAUUUU---CAUUUAUGC----------------CGGGGCGUUAUUUAAUUUAUAUAACGUUCUUAUUUUGAUGGGCAUUUUCGCUGAAUGAACACGA .(((((.((((...(((-(((((.....---)))).....----------------.((((((((((.........))))))))))...........))))...)))).)))))...... ( -22.80) >DroAna_CAF1 224531 119 - 1 CUCGUUAAGUGUCUGUA-AAAUUGUUUUGGGCCUCUAUACUACACAAGUCUGUGGAUGGUGCGUUAUUUGAUUUAUACGUCGUUUCCAUUUUGUGUGGCAUUUUGUCUGAAUGAACACGA ........((((..((.-........(..(((....((.(((((((((...(((((....((((............))))....)))))))))))))).))...)))..)))..)))).. ( -24.40) >consensus CUCGUUAAGUGCCUGCG_AAAUGAUUUU___CAUUUAUGC________________CGGGGCGUUAUUUGAUUUAUAUAACGCUCUUAUUUUGAUGGGCAUUUUCUCUGAAUGAACACGA .((((((((((((.(((.(((((........))))).))).................((((((((((.........))))))))))..........)))))))......)))))...... (-17.86 = -17.92 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:45 2006