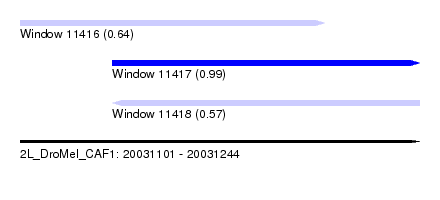
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,031,101 – 20,031,244 |
| Length | 143 |
| Max. P | 0.987232 |

| Location | 20,031,101 – 20,031,210 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -14.32 |
| Energy contribution | -15.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031101 109 + 22407834 CAGCAGAGCUGGCGGAAGAUGUUUCAAUUAUAU----UCUCAUUAAUUUUCAAACAUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGGAAUAAA ((((...)))).((((((((((((.(((((...----......)))))...))))))))))....(((((((((.((((.((....)).)))).))))))))).))....... ( -27.90) >DroSec_CAF1 213503 109 + 1 CAGCAGAGCUGGCGGAAGAUGUUUCAAUUACAU----UCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUAAAAUGAAAUCGCAAUACA ((((...))))(((((..((((.......))))----..))........................((((((((..((((.((....)).))))..)))))))).)))...... ( -26.40) >DroSim_CAF1 210837 109 + 1 CAGCAGAGCUGGCGGAAGAUGUUUCAAUUAUAU----UCUCAUUAAUUAACAAACUUUUUCAUAUUUUCAUUUGUCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACA ((((...))))((((((((.((((.(((((...----......)))))...)))).)))))....(((((((((.((((.((....)).)))).))))))))).)))...... ( -27.20) >DroEre_CAF1 207988 109 + 1 CAGCAGUGCUUUCGGAAAUUGUUUCAAUUAUGU----UCUCAUUCAAUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAGAAGCUGCGGUUAAAUGAAAUCGCAAUGUA ..(((.(((.....((((..((((.(((((((.----...)))..))))..))))..))))....(((((((((...(((((((...))))))))))))))))..))).))). ( -26.80) >DroYak_CAF1 206842 108 + 1 C---AUUAUUUUCU--CAUUGUUUCAAUUAUGUAUAUUAAAAUUAAUUUUCAAACUUUUUCUUAUUUUUAUUUGGCAGCCGCAAUAGCAGCGGUUAAAUGAAAUCCCAACGUA .---..........--.................................................(((((((((((.((.((....)).)).))))))))))).......... ( -13.20) >consensus CAGCAGAGCUGGCGGAAGAUGUUUCAAUUAUAU____UCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACA ..............(((((.((((.(((((.............)))))...)))).)))))....((((((((..((((.((....)).))))..)))))))).......... (-14.32 = -15.52 + 1.20)

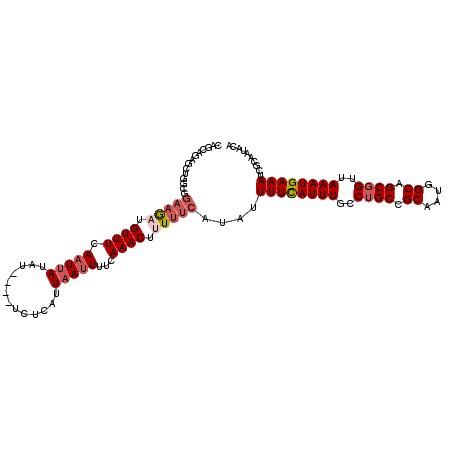

| Location | 20,031,134 – 20,031,244 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -20.26 |
| Energy contribution | -19.98 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031134 110 + 22407834 UCUCAUUAAUUUUCAAACAUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGGAAUAAAUUUUAUUUCCUAAGGCUUCAUGCCUUCGGCUAAA .....................(((....(((((((((.((((.((....)).)))).)))))))))...)))............((.(((((.....))))).))..... ( -25.60) >DroSec_CAF1 213536 110 + 1 UCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUAAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUAAA ....................................(((((((((.......))))))(((((((((........)))))))))....((((.....))))..))).... ( -20.50) >DroSim_CAF1 210870 110 + 1 UCUCAUUAAUUAACAAACUUUUUCAUAUUUUCAUUUGUCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUACA ............................(((((((((.((((.((....)).)))).)))))))))..................((..((((.....))))..))..... ( -21.00) >DroEre_CAF1 208021 108 + 1 UCUCAUUCAAUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAGAAGCUGCGGUUAAAUGAAAUCGCAAUGUAUU-UAUGUUUUUAGGCUU-CUGCCUUCGGGAAAA ((((..........((((.....(((..(((((((((...(((((((...)))))))))))))))).....)))....-...))))..((((..-..))))..))))... ( -25.20) >DroYak_CAF1 206874 110 + 1 UAAAAUUAAUUUUCAAACUUUUUCUUAUUUUUAUUUGGCAGCCGCAAUAGCAGCGGUUAAAUGAAAUCCCAACGUAUUUUACAUCUUUAGGCUUUUUGCCUUCGGGUAAA ............................(((((((((((.((.((....)).)).))))))))))).(((...(((...)))......((((.....))))..))).... ( -22.20) >consensus UCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUAAA ............................((((((((..((((.((....)).))))..))))))))..................((..((((.....))))..))..... (-20.26 = -19.98 + -0.28)


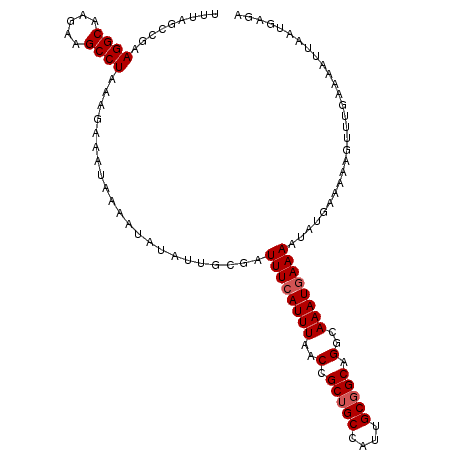
| Location | 20,031,134 – 20,031,244 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20031134 110 - 22407834 UUUAGCCGAAGGCAUGAAGCCUUAGGAAAUAAAAUUUAUUCCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAUGUUUGAAAAUUAAUGAGA ((((.((.(((((.....))))).))...)))).((((((......((((((..(.(((((....))))).)..))))))((((((....)))))).......)))))). ( -26.70) >DroSec_CAF1 213536 110 - 1 UUUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUGUAUUGCGAUUUCAUUUUACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAAGUUUGAAAAUUAAUGAGA ....(((..((((.....)))).......(((((((...........)))))))..(((((....))))).))).................................... ( -20.60) >DroSim_CAF1 210870 110 - 1 UGUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUGUAUUGCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGACAAAUGAAAAUAUGAAAAAGUUUGUUAAUUAAUGAGA .........((((.....)))).....((((((...........((((((((.....(((((.....)))))..))))))))...........))))))........... ( -19.85) >DroEre_CAF1 208021 108 - 1 UUUUCCCGAAGGCAG-AAGCCUAAAAACAUA-AAUACAUUGCGAUUUCAUUUAACCGCAGCUUCUGCGGCAGGCAAAUGAAAAUAUGAAAAAGUUUGAAAUUGAAUGAGA .........((((..-..)))).........-....((((.((((((((.....((((((...))))))....((.((....)).))........))))))))))))... ( -23.90) >DroYak_CAF1 206874 110 - 1 UUUACCCGAAGGCAAAAAGCCUAAAGAUGUAAAAUACGUUGGGAUUUCAUUUAACCGCUGCUAUUGCGGCUGCCAAAUAAAAAUAAGAAAAAGUUUGAAAAUUAAUUUUA ....(((..((((.....))))...((((((...))))))))).((((........(((((....)))))..........((((........)))))))).......... ( -21.10) >consensus UUUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUAUAUUGCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAAGUUUGAAAAUUAAUGAGA .........((((.....))))......................((((((((..(.(((((....))))).)..))))))))............................ (-17.40 = -17.80 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:43 2006