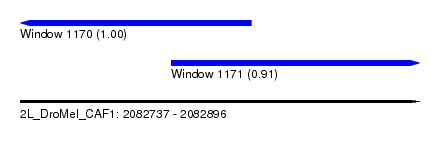
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,082,737 – 2,082,896 |
| Length | 159 |
| Max. P | 0.998854 |

| Location | 2,082,737 – 2,082,829 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -34.99 |
| Energy contribution | -34.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2082737 92 - 22407834 UGGC------UCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACGCACUCGC----ACAC ..((------((.....))))(.(((((((((.((..(((((..((((((((.....))))))))........)))))..)).))))).)))))----.... ( -37.60) >DroSec_CAF1 99873 92 - 1 UGGC------UCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACACACUCGC----ACAC ..((------((.....))))(.(((((((((.((..(((((..((((((((.....))))))))........)))))..)).))))).)))))----.... ( -38.00) >DroSim_CAF1 99662 92 - 1 UGGC------UCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACACACUCGC----ACAC ..((------((.....))))(.(((((((((.((..(((((..((((((((.....))))))))........)))))..)).))))).)))))----.... ( -38.00) >DroEre_CAF1 102309 98 - 1 UGGCUCCGGCUCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACACACUCGC----ACAC ..((....((((.....))))...((((((((.((..(((((..((((((((.....))))))))........)))))..)).))))).)))))----.... ( -39.60) >DroYak_CAF1 103938 92 - 1 UGGC------UCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACACACUCGC----ACAC ..((------((.....))))(.(((((((((.((..(((((..((((((((.....))))))))........)))))..)).))))).)))))----.... ( -38.00) >DroAna_CAF1 99045 96 - 1 UGGU------UUCGUGUGAGUAAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCAGCAAACACACUCGCUCGCACAC ....------...((((((((...((((((((.(((.(((((..((((((((.....))))))))........))))).))).))))).))))))))))).. ( -43.20) >consensus UGGC______UCCGUGUGAGCGAUGAGGUGUUGGUUAGCCUUUGGGCCAGGCAAAUUGCUUGGUCAGUUCUUUGAGGCCGGCAAACACACUCGC____ACAC .............((((..((((....(((((.((..(((((..((((((((.....))))))))........)))))..)).)))))..))))....)))) (-34.99 = -34.88 + -0.11)


| Location | 2,082,797 – 2,082,896 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -22.05 |
| Energy contribution | -24.17 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2082797 99 + 22407834 CUAACCAACACCUCAUCGCUCACACGGA------GCCAAAGAGCCA-AGAGCCAAUGGAUGAGAUAAAACAAUGGUGA-GUGGC-AAGUGGU---G-----AAGUGGAGACGAUGG .............((((((((.(((...------((((....((((-...((((.((..(......)..)).))))..-.))))-...))))---.-----..)))))).))))). ( -34.20) >DroSec_CAF1 99933 102 + 1 CUAACCAACACCUCAUCGCUCACACGGA------GCCAAAGAGCCA-AGAGCCAAUGGGCGAGAUAAAACAAUGGUGA-GUGGC-AAGUGGCGAUG-----GAGUGGAGGCGAUGG .............(((((((..(((...------((((....((((-...((((.((............)).))))..-.))))-...))))....-----..)))..))))))). ( -35.90) >DroSim_CAF1 99722 102 + 1 CUAACCAACACCUCAUCGCUCACACGGA------GCCAAAGAGCCA-AGAGCCAAUGGGCGAGAUAAAACAAUGGUGA-GUGGC-AAGUGGCGAUG-----GAGUGGAGGCGAUGG .............(((((((..(((...------((((....((((-...((((.((............)).))))..-.))))-...))))....-----..)))..))))))). ( -35.90) >DroEre_CAF1 102369 99 + 1 CUAACCAACACCUCAUCGCUCACACGGAGCCGGAGCCAAAGAGCCGAAGAGCCAAUGGGCGAGAUAAAACAAUGGUGA-----------------GUGGCAAAGUGGCGGCAGUGG ....(((...((((...((((.....))))..))((((....((((....((((.((............)).))))..-----------------.))))....))))))...))) ( -28.80) >DroYak_CAF1 103998 109 + 1 CUAACCAACACCUCAUCGCUCACACGGA------GCCAAAGAGCCAAUGGGCCAAUGGGCGAGAUAAAACAAUGGUGAAGUGGUGAAGUGGCGAAGUGGCAAUGUGGC-GAAGUGG ........(((.((..(((...(((...------((((....((((....((((.((............)).))))....))))....))))...))).....)))..-)).))). ( -31.50) >consensus CUAACCAACACCUCAUCGCUCACACGGA______GCCAAAGAGCCA_AGAGCCAAUGGGCGAGAUAAAACAAUGGUGA_GUGGC_AAGUGGCGA_G_____AAGUGGAGGCGAUGG .............(((((((..(((.........((((....((((....((((.((............)).))))....))))....))))...........)))..))))))). (-22.05 = -24.17 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:40 2006