
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,026,966 – 20,027,075 |
| Length | 109 |
| Max. P | 0.655872 |

| Location | 20,026,966 – 20,027,075 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.62 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
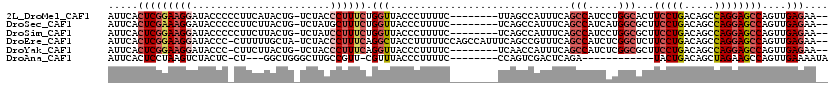
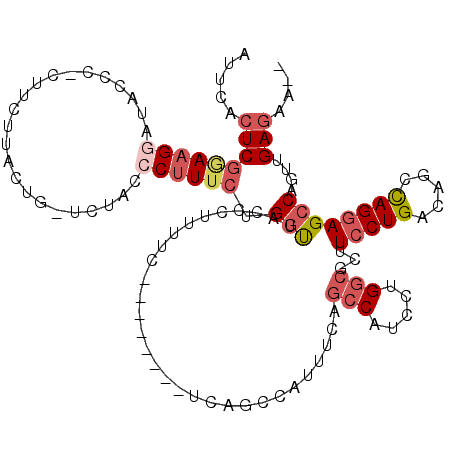
>2L_DroMel_CAF1 20026966 109 - 22407834 AUUCACUCGGAAGGAUACCCCCUUCAUACUG-UCUACCCUUUCUGGUUACCCUUUUC--------UUAGCCAUUUCAGCCAUCCUGGCACUUCCUGACAGCCAGGAGCCAGUUGAGAA-- .........(((((......)))))......-...........((((((........--------.))))))(((((((..(((((((...........)))))))....))))))).-- ( -29.30) >DroSec_CAF1 203820 109 - 1 AUUCACUCGAAAGGAUACCCCCUUCUUACUG-UCUAUGCUUUCUGGUUACCCUUUUC--------UCAGCCAUUUCAGCCAUCAUGGCGCUUCCUGACAGCCAGGAGCCAGUUGAGAA-- ....(((.(((((.(((..(..........)-..))).))))).))).......(((--------(((((.......(((.....)))(((.((((.....)))))))..))))))))-- ( -29.00) >DroSim_CAF1 206539 109 - 1 AUUCACUCGGAAGGAUACCCCCUUCUUACUG-UCUAUCCUUUCUGGUUACCCUUUUC--------UCAGCCAUUUCAGCCAUCCUGGCGCUUCCUGACAGCCAGGAGCCAGUUGAGAA-- ....(((.(((((((((..(..........)-..))))))))).))).......(((--------(((((.......(((.....)))(((.((((.....)))))))..))))))))-- ( -33.20) >DroEre_CAF1 203655 116 - 1 AUUCACUCGGAAGGAUACCC-CUUUUUGCUA-UCUACCCUUUCAGGCUACCUUUUUCCAGCCAUUUCAGCCGUUUCAGCCAUCUCGGCUCUUCCUGACAGCCAGGAGCCAGUUGAGAA-- ........((((((......-..........-.....)))))).((((..........)))).(((((((......((((.....)))).((((((.....))))))...))))))).-- ( -27.40) >DroYak_CAF1 202242 108 - 1 AUUCACUCGGAAGGAUACCC-CUUCUUACUG-UCUACCCUUUCAGGUUACCCUUUUC--------UCAACCAUUUCAGCCAUCUCGGCGCUUCCUGACAGCCAGGAGCCAGUUGAGAA-- ........((((((.....)-)))))....(-(..(((......))).))....(((--------(((((.......(((.....)))(((.((((.....)))))))..))))))))-- ( -30.10) >DroAna_CAF1 220101 95 - 1 AUUCACUCCUAAGUCUACUC-CU---GGCUGGGCUUGCCGUU-CGUUUACCCUUUUC--------CCAGUCGACUCAGA------------UACUGACAGCUAGAAGCCAGUUGAAAAUA .((((...((.((((.....-..---(((((((......((.-.....))......)--------)))))))))).)).------------.((((...((.....)))))))))).... ( -17.71) >consensus AUUCACUCGGAAGGAUACCC_CUUCUUACUG_UCUACCCUUUCUGGUUACCCUUUUC________UCAGCCAUUUCAGCCAUCCUGGCGCUUCCUGACAGCCAGGAGCCAGUUGAGAA__ .....(((((((((.......................)))))).(((..............................(((.....)))...(((((.....))))))))....))).... (-13.30 = -14.62 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:39 2006