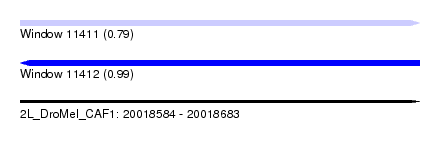
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,018,584 – 20,018,683 |
| Length | 99 |
| Max. P | 0.991613 |

| Location | 20,018,584 – 20,018,683 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -29.66 |
| Energy contribution | -30.97 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20018584 99 + 22407834 GCCCAAAGCACAUCAAUUAUGCAGCUCCGGGCGAACACCAGCGGAGGUGUUUGAGGAAACCACCUGAUGAUGAUGAUGAGGUGGGAGGACGCGUCCGAA ((((..(((.(((.....)))..)))..)))).........((((.((((((.......((((((.((.......)).))))))..)))))).)))).. ( -35.00) >DroSec_CAF1 196097 95 + 1 GCCCGAAGCACAUCAAUUAUGCAGCUCCGGGCGAACACCAGCGGAGGUGUUUGAGGAAACCACCUGAUGA---UGAUGAGGUGGGAGGAC-CGACCGAA (((((.(((.(((.....)))..))).)))))(((((((......)))))))..((...((((((.((..---..)).)))))).....)-)....... ( -35.20) >DroSim_CAF1 198687 95 + 1 GCCCGAAGCACAUCAAUUAUGCAGCUCCGGGCGAACACCAGCGGAGGUGUUUGAGGAAACCACCUGAUGA---UGAUGAGGUGGGAGGAC-CGACCGAA (((((.(((.(((.....)))..))).)))))(((((((......)))))))..((...((((((.((..---..)).)))))).....)-)....... ( -35.20) >DroYak_CAF1 194560 94 + 1 GCCCGAAGCACAUAAAUUAUGCAGCUCCUGGCGAACU-GAGGUGAGGUGUUCGAGGAAACCACCUGACGA---UGAUGAGGUGGGAGGAC-GCUCCGAA (((.(.(((.((((...))))..))).).))).....-..((...(((((((.......((((((.(...---...).))))))..))))-)))))... ( -26.50) >consensus GCCCGAAGCACAUCAAUUAUGCAGCUCCGGGCGAACACCAGCGGAGGUGUUUGAGGAAACCACCUGAUGA___UGAUGAGGUGGGAGGAC_CGACCGAA (((((.(((.(((.....)))..))).)))))(((((((......))))))).......((((((.((.......)).))))))..((......))... (-29.66 = -30.97 + 1.31)

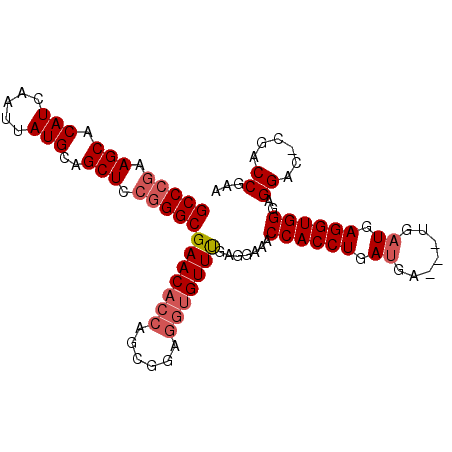
| Location | 20,018,584 – 20,018,683 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.95 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20018584 99 - 22407834 UUCGGACGCGUCCUCCCACCUCAUCAUCAUCAUCAGGUGGUUUCCUCAAACACCUCCGCUGGUGUUCGCCCGGAGCUGCAUAAUUGAUGUGCUUUGGGC ...(((....)))..((((((.((.......)).))))))........((((((......)))))).(((((((((.((((.....))))))))))))) ( -35.10) >DroSec_CAF1 196097 95 - 1 UUCGGUCG-GUCCUCCCACCUCAUCA---UCAUCAGGUGGUUUCCUCAAACACCUCCGCUGGUGUUCGCCCGGAGCUGCAUAAUUGAUGUGCUUCGGGC .......(-(.....((((((.((..---..)).))))))...))...((((((......)))))).(((((((((.((((.....))))))))))))) ( -35.60) >DroSim_CAF1 198687 95 - 1 UUCGGUCG-GUCCUCCCACCUCAUCA---UCAUCAGGUGGUUUCCUCAAACACCUCCGCUGGUGUUCGCCCGGAGCUGCAUAAUUGAUGUGCUUCGGGC .......(-(.....((((((.((..---..)).))))))...))...((((((......)))))).(((((((((.((((.....))))))))))))) ( -35.60) >DroYak_CAF1 194560 94 - 1 UUCGGAGC-GUCCUCCCACCUCAUCA---UCGUCAGGUGGUUUCCUCGAACACCUCACCUC-AGUUCGCCAGGAGCUGCAUAAUUUAUGUGCUUCGGGC .(((((((-......((((((.((..---..)).)))))).((((((((((..........-.)))))..)))))..(((((...)))))))))))).. ( -26.80) >consensus UUCGGACG_GUCCUCCCACCUCAUCA___UCAUCAGGUGGUUUCCUCAAACACCUCCGCUGGUGUUCGCCCGGAGCUGCAUAAUUGAUGUGCUUCGGGC ...((......))..((((((.((.......)).))))))........((((((......)))))).(((((((((.((((.....))))))))))))) (-30.33 = -30.95 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:38 2006